|
Gene Expression Profiling
In the field of molecular biology, gene expression profiling is the measurement of the activity (the gene expression, expression) of thousands of genes at once, to create a global picture of cellular function. These profiles can, for example, distinguish between cells that are actively dividing, or show how the cells react to a particular treatment. Many experiments of this sort measure an entire genome simultaneously, that is, every gene present in a particular cell. Several transcriptomics technologies can be used to generate the necessary data to analyse. DNA microarrays measure the relative activity of previously identified target genes. Sequence based techniques, like RNA-Seq, provide information on the sequences of genes in addition to their expression level. Background Expression profiling is a logical next step after genome sequencing, sequencing a genome: the sequence tells us what the cell could possibly do, while the expression profile tells us what it is actually d ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heatmap
A heat map (or heatmap) is a 2-dimensional data visualization technique that represents the magnitude of individual values within a dataset as a color. The variation in color may be by hue or brightness, intensity. In some applications such as crime analytics or website click-tracking, color is used to represent the ''density'' of data points rather than a value associated with each point. "Heat map" is a relatively new term, but the practice of shading matrices has existed for over a century. History Heat maps originated in 2D displays of the values in a data matrix. Larger values were represented by small dark gray or black squares (pixels) and smaller values by lighter squares. The earliest known example dates to 1873, when Toussaint Loua used a hand-drawn and colored shaded matrix to visualize social statistics across the districts of Paris. The idea of reordering rows and columns to reveal structure in a data matrix, known as Seriation (statistics), seriation, was introduc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Posttranslational Modification
In molecular biology, post-translational modification (PTM) is the covalent process of changing proteins following protein biosynthesis. PTMs may involve enzymes or occur spontaneously. Proteins are created by ribosomes, which translate mRNA into polypeptide chains, which may then change to form the mature protein product. PTMs are important components in cell signalling, as for example when prohormones are converted to hormones. Post-translational modifications can occur on the amino acid side chains or at the protein's C- or N- termini. They can expand the chemical set of the 22 amino acids by changing an existing functional group or adding a new one such as phosphate. Phosphorylation is highly effective for controlling the enzyme activity and is the most common change after translation. Many eukaryotic and prokaryotic proteins also have carbohydrate molecules attached to them in a process called glycosylation, which can promote protein folding and improve stability a ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
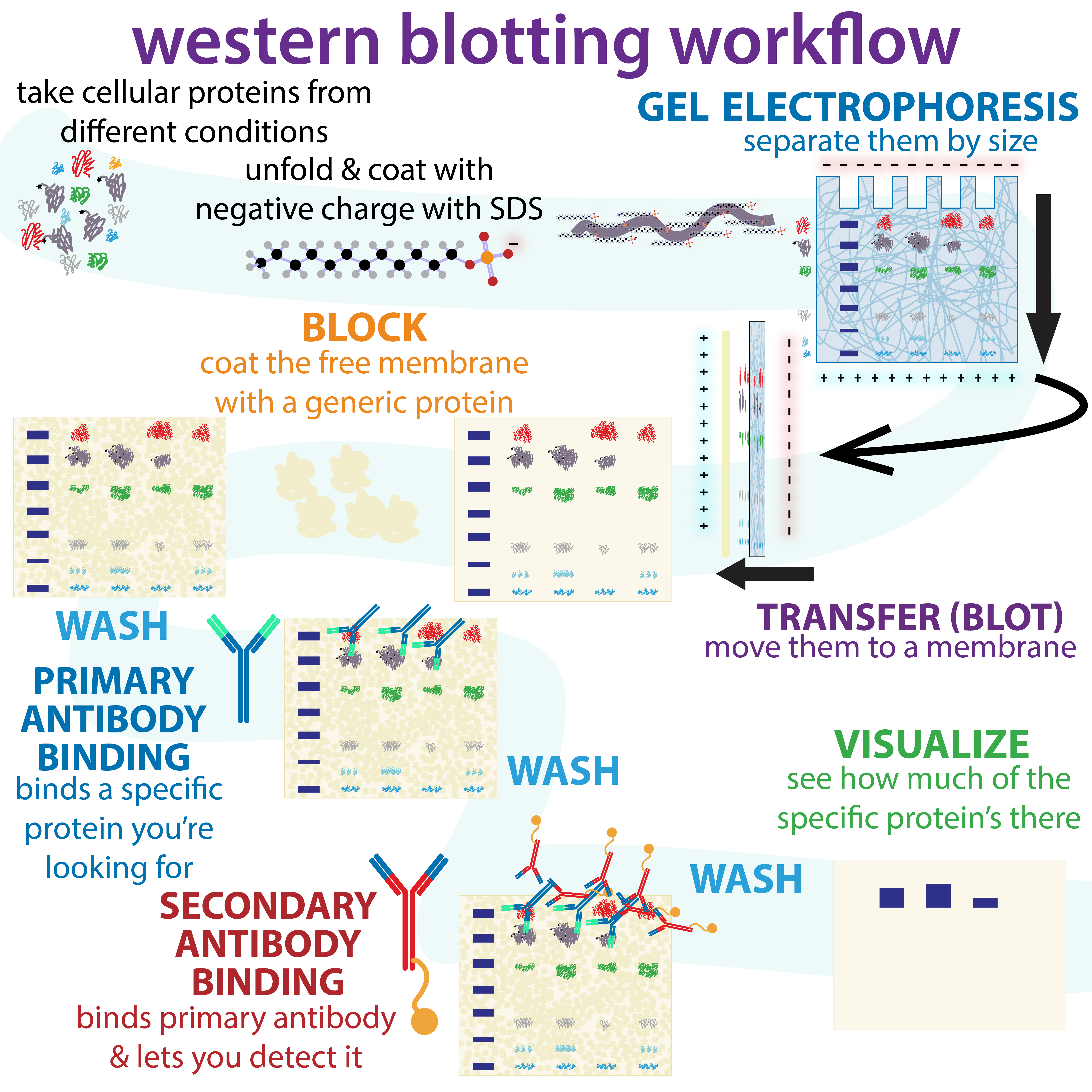
Western Blot
The western blot (sometimes called the protein immunoblot), or western blotting, is a widely used analytical technique in molecular biology and immunogenetics to detect specific proteins in a sample of tissue homogenate or extract. Besides detecting the proteins, this technique is also utilized to visualize, distinguish, and quantify the different proteins in a complicated protein combination. Western blot technique uses three elements to achieve its task of separating a specific protein from a complex: separation by size, transfer of protein to a solid support, and marking target protein using a primary and secondary antibody to visualize. A synthetic or animal-derived antibody (known as the primary antibody) is created that recognizes and binds to a specific target protein. The electrophoresis membrane is washed in a solution containing the primary antibody, before excess antibody is washed off. A secondary antibody is added which recognizes and binds to the primary antibod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Base Pairing
A base pair (bp) is a fundamental unit of double-stranded nucleic acids consisting of two nucleobases bound to each other by hydrogen bonds. They form the building blocks of the DNA double helix and contribute to the folded structure of both DNA and RNA. Dictated by specific hydrogen bonding patterns, "Watson–Crick" (or "Watson–Crick–Franklin") base pairs (guanine–cytosine and adenine–thymine) allow the DNA helix to maintain a regular helical structure that is subtly dependent on its nucleotide sequence. The complementary nature of this based-paired structure provides a redundant copy of the genetic information encoded within each strand of DNA. The regular structure and data redundancy provided by the DNA double helix make DNA well suited to the storage of genetic information, while base-pairing between DNA and incoming nucleotides provides the mechanism through which DNA polymerase replicates DNA and RNA polymerase transcribes DNA into RNA. Many DNA-binding proteins ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
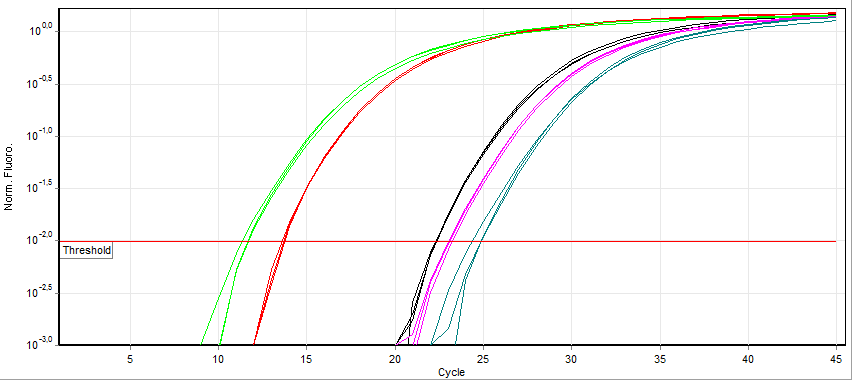
Quantitative PCR
A real-time polymerase chain reaction (real-time PCR, or qPCR when used quantitatively) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively and semi-quantitatively (i.e., above/below a certain amount of DNA molecules). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescent reporter, which permits detection only after hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines propose that the abbreviation ''qPCR'' be used for quantitative real-time PCR and that ' ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray Databases
A microarray database is a repository containing microarray gene expression data. The key uses of a microarray database are to store the measurement data, manage a searchable index, and make the data available to other applications for analysis and interpretation (either directly, or via user downloads). Microarray databases can fall into two distinct classes: # A peer reviewed, public repository that adheres to academic or industry standards and is designed to be used by many analysis applications and groups. A good example of this is the Gene Expression Omnibus (GEO) from NCBI or ArrayExpress from EBI. # A specialized repository associated primarily with the brand of a particular entity (lab, company, university, consortium, group), an application suite, a topic, or an analysis method, whether it is commercial, non-profit, or academic. These databases might have one or more of the following characteristics: #* A subscription or license may be needed to gain full access, #* The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Microarray Analysis Techniques
Microarray analysis techniques are used in interpreting the data generated from experiments on DNA (Gene chip analysis), RNA, and protein microarrays, which allow researchers to investigate the expression state of a large number of genesin many cases, an organism's entire genomein a single experiment. Such experiments can generate very large amounts of data, allowing researchers to assess the overall state of a cell or organism. Data in such large quantities is difficultif not impossibleto analyze without the help of computer programs. Introduction Microarray data analysis is the final step in reading and processing data produced by a microarray chip. Samples undergo various processes including purification and scanning using the microchip, which then produces a large amount of data that requires processing via computer software. It involves several distinct steps, as outlined in the image below. Changing any one of the steps will change the outcome of the analysis, so the MAQC Proj ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Statistical Power
In frequentist statistics, power is the probability of detecting a given effect (if that effect actually exists) using a given test in a given context. In typical use, it is a function of the specific test that is used (including the choice of test statistic and significance level), the sample size (more data tends to provide more power), and the effect size (effects or correlations that are large relative to the variability of the data tend to provide more power). More formally, in the case of a simple hypothesis test with two hypotheses, the power of the test is the probability that the test correctly rejects the null hypothesis (H_0) when the alternative hypothesis (H_1) is true. It is commonly denoted by 1-\beta, where \beta is the probability of making a type II error (a false negative) conditional on there being a true effect or association. Background Statistical testing uses data from samples to assess, or make inferences about, a statistical population. Fo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cellular Differentiation
Cellular differentiation is the process in which a stem cell changes from one type to a differentiated one. Usually, the cell changes to a more specialized type. Differentiation happens multiple times during the development of a multicellular organism as it changes from a simple zygote to a complex system of tissues and cell types. Differentiation continues in adulthood as adult stem cells divide and create fully differentiated daughter cells during tissue repair and during normal cell turnover. Some differentiation occurs in response to antigen exposure. Differentiation dramatically changes a cell's size, shape, membrane potential, metabolic activity, and responsiveness to signals. These changes are largely due to highly controlled modifications in gene expression and are the study of epigenetics. With a few exceptions, cellular differentiation almost never involves a change in the DNA sequence itself. Metabolic composition, however, gets dramatically altered where st ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cross-validation (statistics)
Cross-validation, sometimes called rotation estimation or out-of-sample testing, is any of various similar model validation techniques for assessing how the results of a statistics, statistical analysis will Generalization error, generalize to an independent data set. Cross-validation includes Resampling (statistics), resampling and sample splitting methods that use different portions of the data to test and train a model on different iterations. It is often used in settings where the goal is prediction, and one wants to estimate how accuracy, accurately a predictive modelling, predictive model will perform in practice. It can also be used to assess the quality of a fitted model and the stability of its parameters. In a prediction problem, a model is usually given a dataset of ''known data'' on which training is run (''training dataset''), and a dataset of ''unknown data'' (or ''first seen'' data) against which the model is tested (called the validation set, validation dataset o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Markov Clustering
In statistics, Markov chain Monte Carlo (MCMC) is a class of algorithms used to draw samples from a probability distribution. Given a probability distribution, one can construct a Markov chain whose elements' distribution approximates it – that is, the Markov chain's equilibrium distribution matches the target distribution. The more steps that are included, the more closely the distribution of the sample matches the actual desired distribution. Markov chain Monte Carlo methods are used to study probability distributions that are too complex or too highly dimensional to study with analytic techniques alone. Various algorithms exist for constructing such Markov chains, including the Metropolis–Hastings algorithm. General explanation Markov chain Monte Carlo methods create samples from a continuous random variable, with probability density proportional to a known function. These samples can be used to evaluate an integral over that variable, as its expected value or varianc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hierarchical Clustering
In data mining and statistics, hierarchical clustering (also called hierarchical cluster analysis or HCA) is a method of cluster analysis that seeks to build a hierarchy of clusters. Strategies for hierarchical clustering generally fall into two categories: * Agglomerative: Agglomerative: Agglomerative clustering, often referred to as a "bottom-up" approach, begins with each data point as an individual cluster. At each step, the algorithm merges the two most similar clusters based on a chosen distance metric (e.g., Euclidean distance) and linkage criterion (e.g., single-linkage, complete-linkage). This process continues until all data points are combined into a single cluster or a stopping criterion is met. Agglomerative methods are more commonly used due to their simplicity and computational efficiency for small to medium-sized datasets . * Divisive: Divisive clustering, known as a "top-down" approach, starts with all data points in a single cluster and recursively splits the clu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |