|
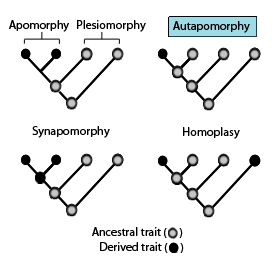
Apomorphies
In phylogenetics, an apomorphy (or derived trait) is a novel character or character state that has evolved from its ancestral form (or plesiomorphy). A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor. ) In cladistics, synapomorphy implies homology. Examples of apomorphy are the presence of erect gait, fur, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a sprawling gait and lack of fur. Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals. Etymology The word —coined by German entomologist Willi Hennig—is derived from the Ancient Greek words (''sún''), meaning "with, together"; (''apó''), meaning "away from"; and (''morphḗ''), meaning "shape, form". Clade analysis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cladistics
Cladistics (; ) is an approach to biological classification in which organisms are categorized in groups (" clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived characteristics ( synapomorphies'')'' that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms ''worms'' or ''fishes'' were used within a ''strict'' cladistic framework, these terms would include humans. Many of these terms are normally used paraphyletically, outside of cladistics, e.g. as a ' grade', which are fruitless to precisely delineate, especially when including extinct species. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cladistics
Cladistics (; ) is an approach to biological classification in which organisms are categorized in groups (" clades") based on hypotheses of most recent common ancestry. The evidence for hypothesized relationships is typically shared derived characteristics ( synapomorphies'')'' that are not present in more distant groups and ancestors. However, from an empirical perspective, common ancestors are inferences based on a cladistic hypothesis of relationships of taxa whose character states can be observed. Theoretically, a last common ancestor and all its descendants constitute a (minimal) clade. Importantly, all descendants stay in their overarching ancestral clade. For example, if the terms ''worms'' or ''fishes'' were used within a ''strict'' cladistic framework, these terms would include humans. Many of these terms are normally used paraphyletically, outside of cladistics, e.g. as a ' grade', which are fruitless to precisely delineate, especially when including extinct species. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cladogram
A cladogram (from Greek ''clados'' "branch" and ''gramma'' "character") is a diagram used in cladistics to show relations among organisms. A cladogram is not, however, an evolutionary tree because it does not show how ancestors are related to descendants, nor does it show how much they have changed, so many differing evolutionary trees can be consistent with the same cladogram. A cladogram uses lines that branch off in different directions ending at a clade, a group of organisms with a last common ancestor. There are many shapes of cladograms but they all have lines that branch off from other lines. The lines can be traced back to where they branch off. These branching off points represent a hypothetical ancestor (not an actual entity) which can be inferred to exhibit the traits shared among the terminal taxa above it. This hypothetical ancestor might then provide clues about the order of evolution of various features, adaptation, and other evolutionary narratives about an ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autapomorphy
In phylogenetics, an autapomorphy is a distinctive feature, known as a derived trait, that is unique to a given taxon. That is, it is found only in one taxon, but not found in any others or outgroup taxa, not even those most closely related to the focal taxon (which may be a species, family or in general any clade). It can therefore be considered an apomorphy in relation to a single taxon. The word ''autapomorphy'', first introduced in 1950 by German entomologist Willi Hennig, is derived from the Greek words αὐτός, ''autos'' "self"; ἀπό, ''apo'' "away from"; and μορφή, ''morphḗ'' = "shape". Discussion Because autapomorphies are only present in a single taxon, they do not convey information about relationship. Therefore, autapomorphies are not useful to infer phylogenetic relationships. However, autapomorphy, like synapomorphy and plesiomorphy is a relative concept depending on the taxon in question. An autapomorphy at a given level may well be a synapomorp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Autapomorphy
In phylogenetics, an autapomorphy is a distinctive feature, known as a derived trait, that is unique to a given taxon. That is, it is found only in one taxon, but not found in any others or outgroup taxa, not even those most closely related to the focal taxon (which may be a species, family or in general any clade). It can therefore be considered an apomorphy in relation to a single taxon. The word ''autapomorphy'', first introduced in 1950 by German entomologist Willi Hennig, is derived from the Greek words αὐτός, ''autos'' "self"; ἀπό, ''apo'' "away from"; and μορφή, ''morphḗ'' = "shape". Discussion Because autapomorphies are only present in a single taxon, they do not convey information about relationship. Therefore, autapomorphies are not useful to infer phylogenetic relationships. However, autapomorphy, like synapomorphy and plesiomorphy is a relative concept depending on the taxon in question. An autapomorphy at a given level may well be a synapomorp ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Synapomorphy
In phylogenetics, an apomorphy (or derived trait) is a novel character or character state that has evolved from its ancestral form (or plesiomorphy). A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor. ) In cladistics, synapomorphy implies homology. Examples of apomorphy are the presence of erect gait, fur, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a sprawling gait and lack of fur. Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals. Etymology The word —coined by German entomologist Willi Hennig—is derived from the Ancient Greek words (''sún''), meaning "with, together"; (''apó''), meaning "away from"; and (''morphḗ''), meaning "shape, form". Clade analysis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Apomorph
In phylogenetics, an apomorphy (or derived trait) is a novel character or character state that has evolved from its ancestral form (or plesiomorphy). A synapomorphy is an apomorphy shared by two or more taxa and is therefore hypothesized to have evolved in their most recent common ancestor. ) In cladistics, synapomorphy implies homology. Examples of apomorphy are the presence of erect gait, fur, the evolution of three middle ear bones, and mammary glands in mammals but not in other vertebrate animals such as amphibians or reptiles, which have retained their ancestral traits of a sprawling gait and lack of fur. Thus, these derived traits are also synapomorphies of mammals in general as they are not shared by other vertebrate animals. Etymology The word —coined by German entomologist Willi Hennig—is derived from the Ancient Greek words (''sún''), meaning "with, together"; (''apó''), meaning "away from"; and (''morphḗ''), meaning "shape, form". Clade analysis ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Tetrapod
Tetrapods (; ) are four-limbed vertebrate animals constituting the superclass Tetrapoda (). It includes extant and extinct amphibians, sauropsids ( reptiles, including dinosaurs and therefore birds) and synapsids ( pelycosaurs, extinct therapsids and all extant mammals). Tetrapods evolved from a clade of primitive semiaquatic animals known as the Tetrapodomorpha which, in turn, evolved from ancient lobe-finned fish (sarcopterygians) around 390 million years ago in the Middle Devonian period; their forms were transitional between lobe-finned fishes and true four-limbed tetrapods. Limbed vertebrates (tetrapods in the broad sense of the word) are first known from Middle Devonian trackways, and body fossils became common near the end of the Late Devonian but these were all aquatic. The first crown-tetrapods ( last common ancestors of extant tetrapods capable of terrestrial locomotion) appeared by the very early Carboniferous, 350 million years ago. The specific aquat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Phylogenetics
In biology, phylogenetics (; from Greek φυλή/ φῦλον [] "tribe, clan, race", and wikt:γενετικός, γενετικός [] "origin, source, birth") is the study of the evolutionary history and relationships among or within groups of organisms. These relationships are determined by Computational phylogenetics, phylogenetic inference methods that focus on observed heritable traits, such as DNA sequences, protein amino acid sequences, or morphology. The result of such an analysis is a phylogenetic tree—a diagram containing a hypothesis of relationships that reflects the evolutionary history of a group of organisms. The tips of a phylogenetic tree can be living taxa or fossils, and represent the "end" or the present time in an evolutionary lineage. A phylogenetic diagram can be rooted or unrooted. A rooted tree diagram indicates the hypothetical common ancestor of the tree. An unrooted tree diagram (a network) makes no assumption about the ancestral line, and do ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homology (biology)
In biology, homology is similarity due to shared ancestry between a pair of structures or genes in different taxa. A common example of homologous structures is the forelimbs of vertebrates, where the Bat wing development, wings of bats and Origin of avian flight, birds, the arms of primates, the front flippers of whales and the forelegs of Quadrupedalism, four-legged vertebrates like Canidae, dogs and crocodiles are all derived from the same ancestral tetrapod structure. Evolutionary biology explains homologous structures adaptation (biology), adapted to different purposes as the result of descent with modification from a Common descent, common ancestor. The term was first applied to biology in a non-evolutionary context by the anatomist Richard Owen in 1843. Homology was later explained by Charles Darwin's theory of evolution in 1859, but had been observed before this, from Aristotle's biology, Aristotle onwards, and it was explicitly analysed by Pierre Belon in 1555. In devel ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Symplesiomorphy
In phylogenetics, a plesiomorphy ("near form") and symplesiomorphy are synonyms for an ancestral character shared by all members of a clade, which does not distinguish the clade from other clades. Plesiomorphy, symplesiomorphy, apomorphy, and synapomorphy, all mean a trait shared between species because they share an ancestral species. Apomorphic and synapomorphic characteristics convey much information about evolutionary clades and can be used to define taxa. However, plesiomorphic and symplesiomorphic characteristics cannot. The term ''symplesiomorphy'' was introduced in 1950 by German entomologist Willi Hennig. Examples A backbone is a plesiomorphic trait shared by birds and mammals, and does not help in placing an animal in one or the other of these two clades. Birds and mammals share this trait because both clades are descended from the same far distant ancestor. Other clades, e.g. snakes, lizards, turtles, fish, frogs, all have backbones and none are either birds nor ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Reptile
Reptiles, as most commonly defined are the animals in the class Reptilia ( ), a paraphyletic grouping comprising all sauropsids except birds. Living reptiles comprise turtles, crocodilians, squamates ( lizards and snakes) and rhynchocephalians ( tuatara). As of March 2022, the Reptile Database includes about 11,700 species. In the traditional Linnaean classification system, birds are considered a separate class to reptiles. However, crocodilians are more closely related to birds than they are to other living reptiles, and so modern cladistic classification systems include birds within Reptilia, redefining the term as a clade. Other cladistic definitions abandon the term reptile altogether in favor of the clade Sauropsida, which refers to all amniotes more closely related to modern reptiles than to mammals. The study of the traditional reptile orders, historically combined with that of modern amphibians, is called herpetology. The earliest known proto-reptiles originated ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |