|
TIM Barrel
The TIM barrel (triose-phosphate isomerase), also known as an alpha/beta barrel, is a conserved protein fold consisting of eight alpha helices (α-helices) and eight parallel beta strands (β-strands) that alternate along the peptide backbone. The structure is named after triose-phosphate isomerase, a conserved metabolic enzyme. TIM barrels are ubiquitous, with approximately 10% of all enzymes adopting this fold. Further, five of seven enzyme commission (EC) enzyme classes include TIM barrel proteins. The TIM barrel fold is evolutionarily ancient, with many of its members possessing little similarity today, instead falling within the ''twilight zone'' of sequence similarity. The inner beta barrel (β-barrel) is in many cases stabilized by intricate salt-bridge networks. Loops at the C-terminal ends of the β-barrel are responsible for catalytic activity while N-terminal end loops are important for the stability of the TIM-barrels. Structural inserts ranging from extend ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Triosephosphateisomerase
Triose-phosphate isomerase (TPI or TIM) is an enzyme () that catalyzes the reversible interconversion of the triose phosphate isomers dihydroxyacetone phosphate and D-glyceraldehyde 3-phosphate. TPI plays an important role in glycolysis and is essential for efficient energy production. TPI has been found in nearly every organism searched for the enzyme, including animals such as mammals and insects as well as in fungi, plants, and bacteria. However, some bacteria that do not perform glycolysis, like ureaplasmas, lack TPI. In humans, deficiencies in TPI are associated with a progressive, severe neurological disorder called triose phosphate isomerase deficiency. Triose phosphate isomerase deficiency is characterized by chronic hemolytic anemia. While there are various mutations that cause this disease, most include the replacement of glutamic acid at position 104 with an aspartic acid. Triose phosphate isomerase is a highly efficient enzyme, performing the reaction billi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's polypeptide chain that is self-stabilizing and that folds independently from the rest. Each domain forms a compact folded three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or disulfide bridges. Domains often form functional units, such as the calcium-binding EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimeric proteins. Background The concept of the domain was first proposed in 1973 by Wetlaufer af ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Indole-3-glycerol-phosphate Synthase
The enzyme indole-3-glycerol-phosphate synthase (IGPS) () catalyzes the chemical reaction :1-(2-carboxyphenylamino)-1-deoxy-D-ribulose 5-phosphate \rightleftharpoons 1-C-(indol-3-yl)-glycerol 3-phosphate + CO2 + H2O This enzyme belongs to the family of lyases, to be specific, the carboxy-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose-5-phosphate carboxy-lyase yclizing 1-C-(indol-3-yl)glycerol-3-phosphate-forming''. Other names in common use include indoleglycerol phosphate synthetase, indoleglycerol phosphate synthase, indole-3-glycerophosphate synthase, 1-(2-carboxyphenylamino)-1-deoxy-D-ribulose-5-phosphate, and carboxy-lyase (cyclizing). This enzyme participates in phenylalanine, tyrosine and tryptophan biosynthesis and two-component system - general. It employs one cofactor, pyruvate. Structural studies In some bacteria, IGPS is a single chain enzyme. In others, such as ''Escherichia coli ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Deoxyribose-phosphate Aldolase
The enzyme deoxyribose-phosphate aldolase () catalyzes the reversible chemical reaction :2-deoxy-D-ribose 5-phosphate \rightleftharpoons D-glyceraldehyde 3-phosphate + acetaldehyde This enzyme belongs to the family of lyases, specifically the aldehyde-lyases, which cleave carbon-carbon bonds. The systematic name of this enzyme class is 2-deoxy-D-ribose-5-phosphate acetaldehyde-lyase (D-glyceraldehyde-3-phosphate-forming). Other names in common use include phosphodeoxyriboaldolase, deoxyriboaldolase, deoxyribose-5-phosphate aldolase, 2-deoxyribose-5-phosphate aldolase, and 2-deoxy-D-ribose-5-phosphate acetaldehyde-lyase. Enzyme Mechanism Amongst aldolases, DERA is unique as it is the only enzyme to afford two aldehydes as products. Crystallography shows that the enzyme is a Class I aldolase, so the mechanism proceeds via the formation of a Schiff base with Lys167 at the active site. A nearby residue, Lys201, is critical to reaction by increasing the acidity of protonated Ly ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
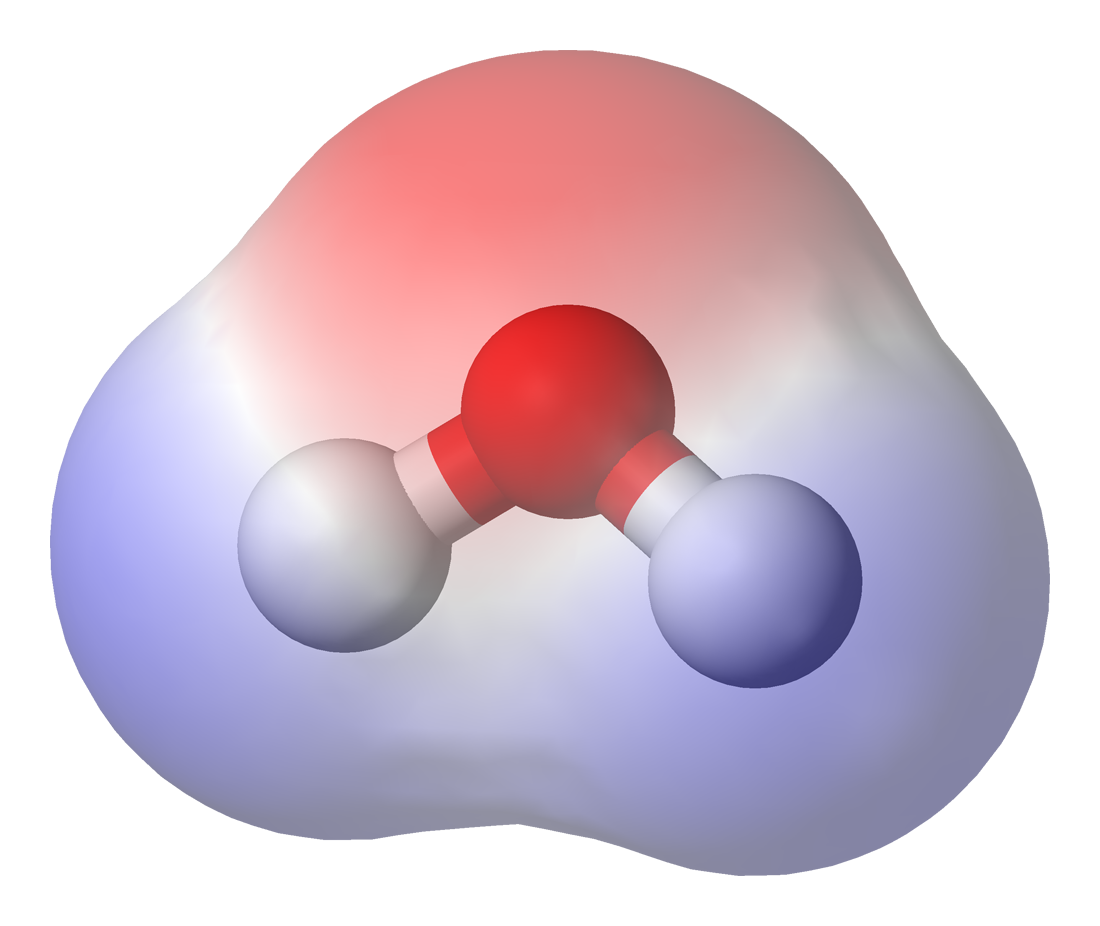
Chemical Polarity
In chemistry, polarity is a separation of electric charge leading to a molecule or its chemical groups having an electric dipole moment, with a negatively charged end and a positively charged end. Polar molecules must contain one or more polar chemical bond, bonds due to a difference in electronegativity between the bonded atoms. Molecules containing polar bonds have no molecular polarity if the Bond dipole moment, bond dipoles cancel each other out by symmetry. Polar molecules interact through dipole–dipole intermolecular forces and hydrogen bonds. Polarity underlies a number of physical properties including surface tension, solubility, and melting and boiling points. Polarity of bonds Not all atoms attract electrons with the same force. The amount of "pull" an atom exerts on its electrons is called its electronegativity. Atoms with high electronegativitiessuch as fluorine, oxygen, and nitrogenexert a greater pull on electrons than atoms with lower electronegativities su ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Residue Depth
Residue depth (RD) is a solvent exposure measure that describes to what extent a residue is buried in the protein structure space. It complements the information provided by conventional accessible surface area (ASA). Currently, predictions in regards to whether a residue is exposed or buried are used in a wide variety of protein structure prediction engines. Such prediction can provide valuable information for protein fold recognition, functional residue prediction and protein drug design. Several biophysical properties of proteins have been shown to correlate with residue depth, including mutant protein stability, protein-protein interface hot-spot, H/D exchange rate of residue and residue conservation. Residue depth has also been utilized in predicting small molecule binding site on proteins, with accuracy statistically on par with other conventional methods. The method has the advantageous of being simple and intuitive. The method has been reported to detect unconventional flat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Shear Number
In protein structures, a beta barrel is a beta sheet composed of tandem repeats that twists and coils to form a closed toroidal structure in which the first strand is bonded to the last strand (hydrogen bond). Beta-strands in many beta-barrels are arranged in an antiparallel fashion. Beta barrel structures are named for resemblance to the barrels used to contain liquids. Most of them are water-soluble proteins and frequently bind hydrophobic ligands in the barrel center, as in lipocalins. Others span cell membranes and are commonly found in porins. Porin-like barrel structures are encoded by as many as 2–3% of the genes in Gram-negative bacteria. It has been shown that more than 600 proteins with various function (e.g., oxidase, dismutase, amylase) contain the beta barrel structure. In many cases, the strands contain alternating polar and non-polar (hydrophilic and hydrophobic) amino acids, so that the hydrophobic residues are oriented into the interior of the barrel to form a h ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Crystallization
Protein crystallization is the process of formation of a regular array of individual protein molecules stabilized by crystal contacts. If the crystal is sufficiently ordered, it will diffract. Some proteins naturally form crystalline arrays, like aquaporin in the lens of the eye. In the process of protein crystallization, proteins are dissolved in an aqueous environment and sample solution until they reach the supersaturated state. Different methods are used to reach that state such as vapor diffusion, microbatch, microdialysis, and free-interface diffusion. Developing protein crystals is a difficult process influenced by many factors, including pH, temperature, ionic strength in the crystallization solution, and even gravity. Once formed, these crystals can be used in structural biology to study the molecular structure of the protein, particularly for various industrial or medical purposes. Development of protein crystallization For over 150 years, scientists from all arou ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Design
Protein design is the rational design of new protein molecules to design novel activity, behavior, or purpose, and to advance basic understanding of protein function. Proteins can be designed from scratch (''de novo'' design) or by making calculated variants of a known protein structure and its sequence (termed ''protein redesign''). Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis. Rational protein design dates back to the mid-1970s. Recently, however, there were numerous examples of successful rational design of water-soluble and even transmembrane peptides and proteins, in part due to a better understanding of different factors contributing to protein structure stability and development of better computational methods. Overview and history The goal in rat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |