|
Toehold Mediated Strand Displacement
Toehold mediated strand displacement (TMSD) is an enzyme-free molecular tool to exchange one strand of DNA or RNA (output) with another strand (input). It is based on the hybridization of two complementary strands of DNA or RNA via Watson-Crick base pairing (A-T/U and C-G) and makes use of a process called branch migration. Although branch migration has been known to the scientific community since the 1970s, TMSD has not been introduced to the field of DNA nanotechnology until 2000 when Yurke et al. was the first who took advantage of TMSD. He used the technique to open and close a set of DNA tweezers made of two DNA helices using an auxiliary strand of DNA as fuel. Since its first use, the technique has been modified for the construction of autonomous molecular motors, catalytic amplifiers, reprogrammable DNA nanostructures and molecular logic gates. It has also been used in conjunction with RNA for the production of kinetically-controlled ribosensors. TMSD starts with a double-stra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Branch Migration
Branch migration is the process by which base pairs on homologous DNA strands are consecutively exchanged at a Holliday junction, moving the branch point up or down the DNA sequence. Branch migration is the second step of genetic recombination, following the exchange of two single strands of DNA between two homologous chromosomes. The process is random, and the branch point can be displaced in either direction on the strand, influencing the degree of which the genetic material is exchanged. Branch migration can also be seen in DNA repair and replication, when filling in gaps in the sequence. It can also be seen when a foreign piece of DNA invades the strand. Mechanism The mechanism for branch migration differs between prokaryotes and eukaryotes. Prokaryotes The mechanism for prokaryotic branch migration has been studied many times in '' Escherichia coli''. In ''E. coli,'' the proteins RuvA and RuvB come together and form a complex that facilitates the process in a nu ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
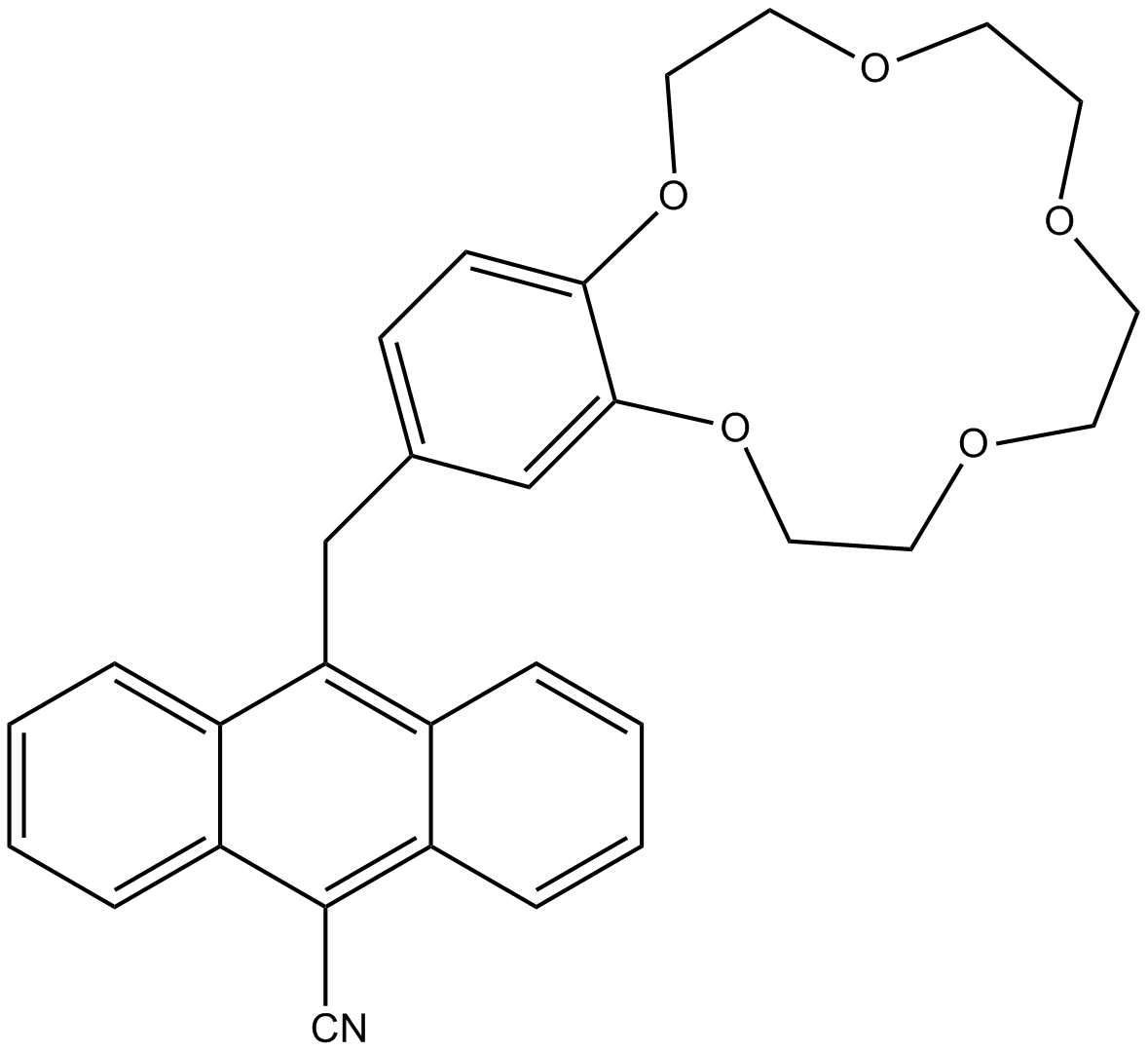
Molecular Logic Gate
A molecular logic gate is a molecule that performs a logical operation based on one or more physical or chemical inputs and a single output. The field has advanced from simple logic systems based on a single chemical or physical input to molecules capable of combinatorial and sequential operations such as arithmetic operations (i.e. moleculators and memory storage algorithms). For logic gates with a single input, there are four possible output patterns. When the input is 0, the output can be either a 0 or 1. When the input is 1, the output can again be 0 or 1. The four output bit patterns that can arise corresponds to a specific logic type: PASS 0, YES, NOT and PASS 1. PASS 0 always outputs 0, whatever the input. PASS 1 always outputs 1, whatever the input. YES outputs a 1 when the input is 1 and NOT is the inverse YES – it outputs a 0 when the input is 1. An example of a YES logic gate is the molecular structure shown below. A "1" output is given only when sodium ions are presen ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Nanotechnology
DNA nanotechnology is the design and manufacture of artificial nucleic acid structures for technological uses. In this field, nucleic acids are used as non-biological engineering materials for nanotechnology rather than as the carriers of genetic information in living Cell (biology), cells. Researchers in the field have created static structures such as two- and three-dimensional Crystal structure, crystal lattices, nanotubes, Polyhedron, polyhedra, and arbitrary shapes, and functional devices such as molecular machines and DNA computing, DNA computers. The field is beginning to be used as a tool to solve basic science problems in structural biology and biophysics, including applications in X-ray crystallography and nuclear magnetic resonance spectroscopy of proteins to determine structures. Potential applications in molecular scale electronics and nanomedicine are also being investigated. The conceptual foundation for DNA nanotechnology was first laid out by Nadrian Seeman in th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase
A polymerase is an enzyme ( EC 2.7.7.6/7/19/48/49) that synthesizes long chains of polymers or nucleic acids. DNA polymerase and RNA polymerase are used to assemble DNA and RNA molecules, respectively, by copying a DNA template strand using base-pairing interactions or RNA by half ladder replication. A DNA polymerase from the thermophilic bacterium, '' Thermus aquaticus'' (''Taq'') ( PDBbr>1BGX EC 2.7.7.7) is used in the polymerase chain reaction, an important technique of molecular biology. A polymerase may be template dependent or template independent. Poly-A-polymerase is an example of template independent polymerase. Terminal deoxynucleotidyl transferase also known to have template independent and template dependent activities. Types By function * DNA polymerase (DNA-directed DNA polymerase, DdDP) **Family A: DNA polymerase I; Pol γ, θ, ν **Family B: DNA polymerase II; Pol α, δ, ε, ζ **Family C: DNA polymerase III holoenzyme **Family X: Pol β, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Toehold Mediated Strand Displacement
Toehold mediated strand displacement (TMSD) is an enzyme-free molecular tool to exchange one strand of DNA or RNA (output) with another strand (input). It is based on the hybridization of two complementary strands of DNA or RNA via Watson-Crick base pairing (A-T/U and C-G) and makes use of a process called branch migration. Although branch migration has been known to the scientific community since the 1970s, TMSD has not been introduced to the field of DNA nanotechnology until 2000 when Yurke et al. was the first who took advantage of TMSD. He used the technique to open and close a set of DNA tweezers made of two DNA helices using an auxiliary strand of DNA as fuel. Since its first use, the technique has been modified for the construction of autonomous molecular motors, catalytic amplifiers, reprogrammable DNA nanostructures and molecular logic gates. It has also been used in conjunction with RNA for the production of kinetically-controlled ribosensors. TMSD starts with a double-stra ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Stem-loop
Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many RNA secondary structures. As an important secondary structure of RNA, it can direct RNA folding, protect structural stability for messenger RNA (mRNA), provide recognition sites for RNA binding proteins, and serve as a substrate for enzymatic reactions. Formation and stability The formation of a stem-loop structure is dependent on the stability of the resulting helix and loop regions. The first prerequisite is the presence of a sequence that can fold back on itself to form a paired double helix. The stability of this helix is determined by its length, the number of mismatches or bulges i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |