|
TET-assisted Pyridine Borane Sequencing
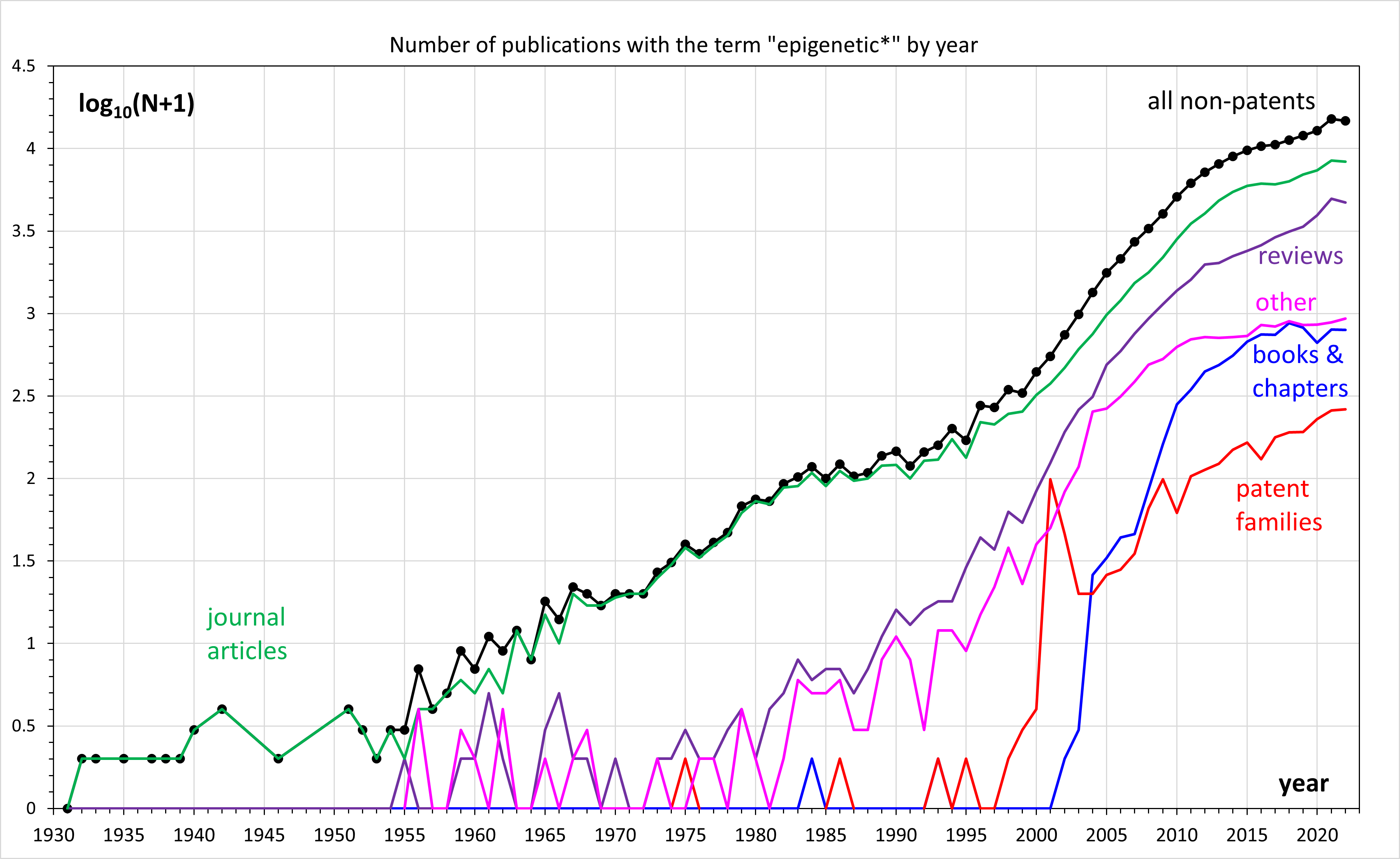
TET-assisted pyridine borane sequencing or TAPS is a laboratory technique in epigenetics for high-throughput profiling of DNA methylation at a single base-pair resolution. It uses a two-step enzymatic conversion of methylated cytosines, 5mC and 5hmC, to uracil which is read as a thymine after sequencing. Due to direct conversion of methylated bases, TAPS is a mC-to-T chemistry unlike traditional method such as bisulfite sequencing or EM-Seq which are C-to-T chemistries and convert un-methylated cytosines. The enzymatic conversion mitigates DNA damage and degradation, similar to EM-Seq, and direct conversion of methylated cytosines without affecting unmodified cytosines further improves the sensitivity and specificity of DNA methylation profiling. TAPS was developed by Chunxiao Song and Benjamin Schuster-Böckler and their groups affiliated with Ludwig Cancer Research at the University of Oxford in 2019 and published in Nature Biotechnology. TAPS is patented in US and its technolog ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Dihydrouracil
Dihydrouracil is an intermediate in the catabolism of uracil. It is the base present in the nucleoside dihydrouridine. See also * Dihydrouracil dehydrogenase (NAD+) * Dihydrouracil oxidase * Dihydropyrimidinase In enzymology, a dihydropyrimidinase () is an enzyme that catalyzes the chemical reaction :5,6-dihydrouracil + H2O \rightleftharpoons 3-ureidopropanoate Thus, the two substrates of this enzyme are 5,6-dihydrouracil and H2O, whereas its produ ... References Nucleobases Ureas Pyrimidinediones {{organic-compound-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Epigenetics
In biology, epigenetics is the study of changes in gene expression that happen without changes to the DNA sequence. The Greek prefix ''epi-'' (ἐπι- "over, outside of, around") in ''epigenetics'' implies features that are "on top of" or "in addition to" the traditional (DNA sequence based) genetic mechanism of inheritance. Epigenetics usually involves a change that is not erased by cell division, and affects the regulation of gene expression. Such effects on cellular and physiological traits may result from environmental factors, or be part of normal development. The term also refers to the mechanism of changes: functionally relevant alterations to the genome that do not involve mutation of the nucleotide sequence. Examples of mechanisms that produce such changes are DNA methylation and histone modification, each of which alters how genes are expressed without altering the underlying DNA sequence. Further, non-coding RNA sequences have been shown to play a key role in the r ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5-hydroxymethylcytosine
5-Hydroxymethylcytosine (5hmC) is a DNA pyrimidine nitrogen base derived from cytosine. It is potentially important in epigenetics, because the hydroxymethyl group on the cytosine can possibly switch a gene on and off. It was first seen in bacteriophages in 1952. However, in 2009 it was found to be abundant in human and mouse brains, as well as in embryonic stem cells. In mammals, it can be generated by oxidation of 5-methylcytosine, a reaction mediated by TET enzymes. Localization Every mammalian cell seems to contain 5-Hydroxymethylcytosine, but the levels vary significantly depending on the cell type. The highest levels are found in neuronal cells of the central nervous system. The amount of hydroxymethylcytosine increases with age, as shown in mouse hippocampus and cerebellum. Function The exact function of this nitrogen base is still not fully elucidated, but it is thought that it may regulate gene expression or prompt DNA demethylation. This hypothesis is supported by th ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5-methylcytosine
5-Methylcytosine (5mC) is a methylation, methylated form of the DNA base cytosine (C) that regulates gene Transcription (genetics), transcription and takes several other biological roles. When cytosine is methylated, the DNA maintains the same sequence, but the Gene expression#DNA methylation and demethylation in transcriptional regulation, expression of methylated genes can be altered (the study of this is part of the field of epigenetics). 5-Methylcytosine is incorporated in the nucleoside 5-Methylcytidine, 5-methylcytidine. Discovery While trying to isolate the bacterial toxin responsible for tuberculosis, W.G. Ruppel isolated a novel nucleic acid named tuberculinic acid in 1898 from ''Mycobacterium tuberculosis, Tubercle bacillus''. The nucleic acid was found to be unusual, in that it contained in addition to thymine, guanine and cytosine, a methylated nucleotide. In 1925, Treat Baldwin Johnson, Johnson and Coghill successfully detected a minor amount of a methylated cytosi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Glycosyltransferase
Glycosyltransferases (GTFs, Gtfs) are enzymes ( EC 2.4) that establish natural glycosidic linkages. They catalyze the transfer of saccharide moieties from an activated nucleotide sugar (also known as the "glycosyl donor") to a nucleophilic glycosyl acceptor molecule, the nucleophile of which can be oxygen- carbon-, nitrogen-, or sulfur-based. The result of glycosyl transfer can be a carbohydrate, glycoside, oligosaccharide, or a polysaccharide. Some glycosyltransferases catalyse transfer to inorganic phosphate or water. Glycosyl transfer can also occur to protein residues, usually to tyrosine, serine, or threonine to give O-linked glycoproteins, or to asparagine to give N-linked glycoproteins. Mannosyl groups may be transferred to tryptophan to generate C-mannosyl tryptophan, which is relatively abundant in eukaryotes. Transferases may also use lipids as an acceptor, forming glycolipids, and even use lipid-linked sugar phosphate donors, such as dolichol phosphates in eukar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase
A DNA polymerase is a member of a family of enzymes that catalyze the synthesis of DNA molecules from nucleoside triphosphates, the molecular precursors of DNA. These enzymes are essential for DNA replication and usually work in groups to create two identical DNA duplexes from a single original DNA duplex. During this process, DNA polymerase "reads" the existing DNA strands to create two new strands that match the existing ones. These enzymes catalysis, catalyze the chemical reaction : deoxynucleoside triphosphate + DNAn pyrophosphate + DNAn+1. DNA polymerase adds nucleotides to the Directionality (molecular biology), three prime (3')-end of a DNA strand, one nucleotide at a time. Every time a Cell division, cell divides, DNA polymerases are required to duplicate the cell's DNA, so that a copy of the original DNA molecule can be passed to each daughter cell. In this way, genetic information is passed down from generation to generation. Before replication can take place, an enzy ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
5-formylcytosine
5-Formylcytosine (5fC) is a pyrimidine nitrogen base derived from cytosine. In the context of nucleic acid chemistry and biology, it is regarded as an epigenetic marker. Discovered in 2011 in mammalian embryonic stem cells by Thomas Carell's research group the modified nucleoside was more recently confirmed to be relevant both as an intermediate in the active demethylation pathway and as a standalone epigenetic marker. In mammals, 5fC is formed by oxidation of 5-hydroxymethylcytosine (5hmC) a reaction mediated by TET enzymes. Its molecular formula is C5H5N3O2. Localization Similarly to the related cytosine modifications 5-methylcytosine (5mC) and 5hmC, 5fC is broadly distributed across the mammalian genome, although it is much more rarely occurring. The specific concentration values vary significantly depending on the cell type. 5fC can be aberrantly expressed in distinct sets of tissue that can indicate different tumor onsets and canceration. Functions The exact functions o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Iron
Iron is a chemical element; it has symbol Fe () and atomic number 26. It is a metal that belongs to the first transition series and group 8 of the periodic table. It is, by mass, the most common element on Earth, forming much of Earth's outer and inner core. It is the fourth most abundant element in the Earth's crust, being mainly deposited by meteorites in its metallic state. Extracting usable metal from iron ores requires kilns or furnaces capable of reaching , about 500 °C (900 °F) higher than that required to smelt copper. Humans started to master that process in Eurasia during the 2nd millennium BC and the use of iron tools and weapons began to displace copper alloys – in some regions, only around 1200 BC. That event is considered the transition from the Bronze Age to the Iron Age. In the modern world, iron alloys, such as steel, stainless steel, cast iron and special steels, are by far the most common industrial metals, due to their mechan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PCR Amplification
The polymerase chain reaction (PCR) is a method widely used to make millions to billions of copies of a specific DNA sample rapidly, allowing scientists to amplify a very small sample of DNA (or a part of it) sufficiently to enable detailed study. PCR was invented in 1983 by American biochemist Kary Mullis at Cetus Corporation. Mullis and biochemist Michael Smith, who had developed other essential ways of manipulating DNA, were jointly awarded the Nobel Prize in Chemistry in 1993. PCR is fundamental to many of the procedures used in genetic testing and research, including analysis of ancient samples of DNA and identification of infectious agents. Using PCR, copies of very small amounts of DNA sequences are exponentially amplified in a series of cycles of temperature changes. PCR is now a common and often indispensable technique used in medical laboratory research for a broad variety of applications including biomedical research and forensic science. The majority of PCR met ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
TET Enzymes
The TET enzymes are a family of ten-eleven translocation (TET) 5-Methylcytosine, methylcytosine dioxygenases. They are instrumental in DNA demethylation. 5-Methylcytosine (see first Figure) is a methylation, methylated form of the DNA base cytosine (C) that often regulates gene Transcription (genetics), transcription and has several other functions in the genome. Demethylation by TET enzymes (see second Figure), can alter the regulation of transcription. The TET enzymes catalyze the hydroxylation of DNA 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), and can further catalyse oxidation of 5hmC to 5-formylcytosine (5fC) and then to 5-carboxycytosine (5caC). 5fC and 5caC can be removed from the DNA base sequence by base excision repair and replaced by cytosine in the base sequence. TET enzymes have central roles in DNA demethylation required during embryogenesis, gametogenesis, Epigenetics in learning and memory, memory, learning, addiction and Nociception, pain pe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |