 |
Short Interspersed Element
Retrotransposons (also called Class I transposable elements) are mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged. Through reverse transcription, retrotransposons amplify themselves quickly to become abundant in eukaryotic genomes such as maize (49–78%) and humans (42%). They are only present in eukaryotes but share features with retroviruses such as HIV, for example, discontinuous reverse transcriptase-mediated extrachromosomal recombination. There are two main types of retrotransposons, long terminal repeats (LTRs) and non-long terminal repeats (non-LTRs). Retrotransposons are classified based on sequence and method of transposition. Most retrotransposons in the maize genome are LTR, whereas in humans they are mostly non-LTR. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
 |
Retrotransposons
Retrotransposons (also called Class I transposable elements) are transposable element, mobile elements which move in the host genome by converting their transcribed RNA into DNA through reverse transcription. Thus, they differ from Class II transposable elements, or DNA transposons, in utilizing an RNA intermediate for the transposition and leaving the transposition donor site unchanged. Through reverse transcription, retrotransposons amplify themselves quickly to become abundant in eukaryote, eukaryotic genomes such as maize (49–78%) and humans (42%). They are only present in eukaryotes but share features with retroviruses such as HIV, for example, discontinuous reverse transcriptase-mediated extrachromosomal recombination. There are two main types of retrotransposons, long terminal repeats (LTRs) and non-long terminal repeats (non-LTRs). Retrotransposons are classified based on sequence and method of transposition. Most retrotransposons in the maize genome are LTR, whereas in ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
Retrotransposon Marker
Retrotransposon markers are components of DNA which are used as cladistic markers. They assist in determining the common ancestry, or not, of related taxa. The "presence" of a given retrotransposon in related taxa suggests their orthologous integration, a derived condition acquired via a common ancestry, while the "absence" of particular elements indicates the plesiomorphic condition prior to integration in more distant taxa. The use of presence/absence analyses to reconstruct the systematic biology of mammals depends on the availability of retrotransposons that were actively integrating before the divergence of a particular species. Details The analysis of SINEs – Short INterspersed Elements – LINEs – Long INterspersed Elements – or truncated LTRs – Long Terminal Repeats – as molecular cladistic markers represents a particularly interesting complement to DNA sequence and morphological data. The reason for this is that retrotransposons are assumed to represent pow ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
RetrOryza
RetrOryza is a database of Long terminal repeat-retrotransposons for the rice genome. See also * Long terminal repeat * Retrotransposon * Rice Rice is a cereal grain and in its Domestication, domesticated form is the staple food of over half of the world's population, particularly in Asia and Africa. Rice is the seed of the grass species ''Oryza sativa'' (Asian rice)—or, much l ... References External links * http://retroryza.fr/ Biological databases Molecular genetics Mobile genetic elements Rice {{Biodatabase-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Paleovirology
Paleovirology is the study of viruses that existed in the past but are now extinct. In general, viruses cannot leave behind physical fossils, therefore indirect evidence is used to reconstruct the past. For example, viruses can cause evolution of their hosts, and the signatures of that evolution can be found and interpreted in the present day. Also, some viral genetic fragments which were integrated into germline cells of an ancient organism have been passed down to our time as viral fossils, or endogenous viral elements (EVEs). EVEs that originate from the integration of retroviruses are known as endogenous retroviruses, or ERVs, and most viral fossils are ERVs. They may preserve genetic code from millions of years ago, hence the "fossil" terminology, although no one has detected a virus in mineral fossils. The most surprising viral fossils originate from non-retroviral DNA and RNA viruses. Terminology Although there is no formal classification system for EVEs, they are catego ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Paleogenetics
Paleogenetics is the study of the past through the examination of preserved genetic material from the remains of ancient organisms. Emile Zuckerkandl and Linus Pauling introduced the term in 1963, long before the sequencing of DNA, in reference to the possible reconstruction of the corresponding polypeptide sequences of past organisms. The first sequence of ancient DNA, isolated from a museum specimen of the extinct quagga, was published in 1984 by a team led by Allan Wilson. Paleogeneticists do not recreate actual organisms, but piece together ancient DNA sequences using various analytical methods. Fossils are "the only direct witnesses of extinct species and of evolutionary events" and finding DNA within those fossils exposes tremendously more information about these species, potentially their entire physiology and anatomy. The most ancient DNA sequence to date was reported in February 2021, from the tooth of a Siberian mammoth frozen for over a million years. Applications ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Interspersed Repeat
Interspersed repetitive DNA is found in all eukaryotic genomes. They differ from tandem repeat DNA in that rather than the repeat sequences coming right after one another, they are dispersed throughout the genome and nonadjacent. The sequence that repeats can vary depending on the type of organism, and many other factors. Certain classes of interspersed repeat sequences propagate themselves by RNA mediated Transposable element, transposition; they have been called retrotransposons, and they constitute 25–40% of most mammalian genomes. Some types of interspersed repetitive DNA elements allow new genes to evolve by uncoupling similar DNA sequences from gene conversion during meiosis. Intrachromosomal and interchromosomal gene conversion Gene conversion acts on DNA sequence homology as its substrate. There is no requirement that the sequence homologies lie at the allele, allelic positions on their respective chromosomes or even that the homologies lie on different chromosomes. Gene ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Insertion Sequences
Insertion element (also known as an IS, an insertion sequence element, or an IS element) is a short DNA sequence that acts as a simple transposable element. Insertion sequences have two major characteristics: they are small relative to other transposable elements (generally around 700 to 2500 bp in length) and only code for proteins implicated in the transposition activity (they are thus different from other transposons, which also carry accessory genes such as antibiotic resistance genes). A particular insertion sequence may be named according to the form IS''n'', where ''n'' is a number (e.g. IS''1'', IS''2'', IS''3'', IS''10'', IS''50'', IS''911'', IS''26'' etc.); this is not the only naming scheme used, however. Composition These proteins are usually the transposase which catalyses the enzymatic reaction allowing the IS to move, and also one regulatory protein which either stimulates or inhibits the transposition activity. The coding region in an insertion sequence is usua ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
Genomic Organization
300px, Genome sizes and corresponding composition of six major model organisms as pie charts. The increase in genome size correlates with the vast expansion of noncoding (i.e., intronic, intergenic, and interspersed repeat sequences) and repeat DNA (e.g., satellite, LINEs, short interspersed nuclear element (SINEs), DNA ( Alu sequence), in red) sequences in more complex multicellular organisms. This expansion is accompanied by an increase in the number of epigenetic mechanisms (particularly repressive) that regulate the genome. Expansion of the genome also correlates with an increase in size and complexity of transcription units, except for plants. P = Promoter DNA element. The hereditary material i.e. DNA (deoxyribonucleic acid) of an organism is composed of a sequence of four nucleotides in a specific pattern, which encodes information as a function of their order. Genomic organization refers to the linear order of DNA elements and their division into chromosomes. "Genome organi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
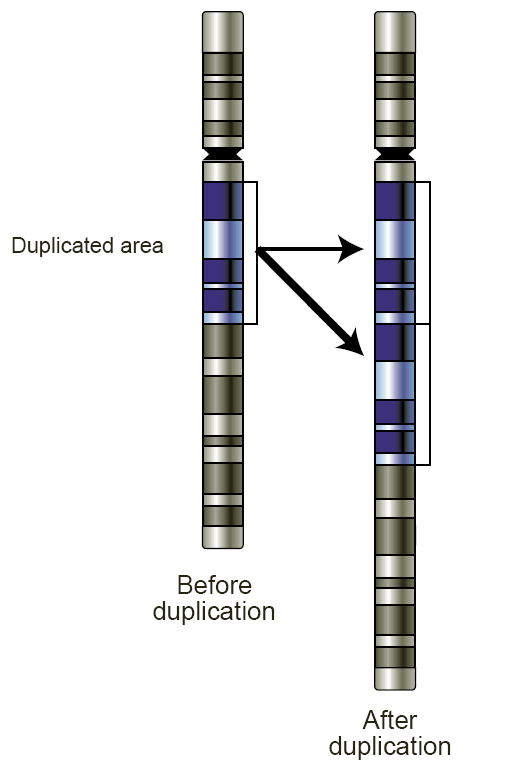 |
Copy-number Variation
Copy number variation (CNV) is a phenomenon in which sections of the genome are repeated and the number of repeats in the genome varies between individuals. Copy number variation is a type of structural variation: specifically, it is a type of duplication or deletion event that affects a considerable number of base pairs. Approximately two-thirds of the entire human genome may be composed of repeats and 4.8–9.5% of the human genome can be classified as copy number variations. In mammals, copy number variations play an important role in generating necessary variation in the population as well as disease phenotype. Copy number variations can be generally categorized into two main groups: short repeats and long repeats. However, there are no clear boundaries between the two groups and the classification depends on the nature of the loci of interest. Short repeats include mainly dinucleotide repeats (two repeating nucleotides e.g. A-C-A-C-A-C...) and trinucleotide repeats. Lon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
Non-coding RNA
A non-coding RNA (ncRNA) is a functional RNA molecule that is not Translation (genetics), translated into a protein. The DNA sequence from which a functional non-coding RNA is transcribed is often called an RNA gene. Abundant and functionally important list of RNAs, types of non-coding RNAs include transfer RNAs (tRNAs) and ribosomal RNAs (rRNAs), as well as small RNAs such as microRNAs, siRNAs, piRNAs, snoRNAs, snRNAs, Extracellular RNA, exRNAs, scaRNAs and the long noncoding RNA, long ncRNAs such as Xist and HOTAIR. The number of non-coding RNAs within the human genome is unknown; however, recent Transcriptomics, transcriptomic and Bioinformatics, bioinformatic studies suggest that there are thousands of non-coding transcripts. Many of the newly identified ncRNAs have unknown functions, if any. There is no consensus on how much of non-coding transcription is functional: some believe most ncRNAs to be non-functional "junk RNA", spurious transcriptions, while others expect that ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |
|
|
RNA Interference
RNA interference (RNAi) is a biological process in which RNA molecules are involved in sequence-specific suppression of gene expression by double-stranded RNA, through translational or transcriptional repression. Historically, RNAi was known by other names, including ''co-suppression'', ''post-transcriptional gene silencing'' (PTGS), and ''quelling''. The detailed study of each of these seemingly different processes elucidated that the identity of these phenomena were all actually RNAi. Andrew Fire and Craig Mello shared the 2006 Nobel Prize in Physiology or Medicine for their work on RNAi in the nematode worm ''Caenorhabditis elegans'', which they published in 1998. Since the discovery of RNAi and its regulatory potentials, it has become evident that RNAi has immense potential in suppression of desired genes. RNAi is now known as precise, efficient, stable and better than antisense therapy for gene suppression. Antisense RNA produced intracellularly by an expression vector may be ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] [Amazon] |