|
Poly ADP Ribose Polymerase
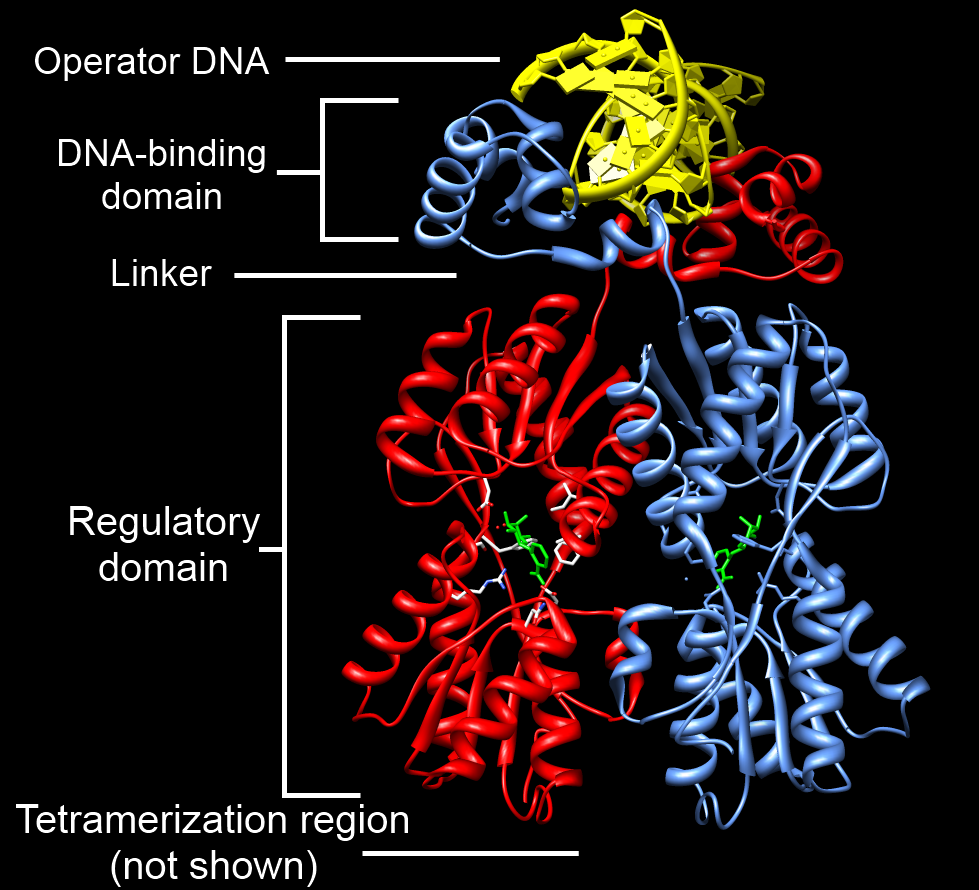
Poly (ADP-ribose) polymerase (PARP) is a family of proteins involved in a number of cellular processes such as DNA repair, genomic stability, and programmed cell death. Members of PARP family The PARP family comprises 17 members (10 putative). They vary greatly in structure and function within the cell. * ''PARP1'', ''PARP2'', VPARP ('' PARP4''), Tankyrase-1 and -2 (PARP-5a or '' TNKS'', and PARP-5b or '' TNKS2'') have a confirmed PARP activity. * Others include '' PARP3'', , '' TIPARP'' (or "PARP7"), '' PARP8'', , '' PARP10'', , ''PARP12'', , , and '' PARP16''. Structure PARP is composed of four domains of interest: a DNA-binding domain, a caspase-cleaved domain (see below), an auto-modification domain, and a catalytic domain. The DNA-binding domain is composed of two zinc finger motifs. In the presence of damaged DNA (base pair-excised), the DNA-binding domain will bind the DNA and induce a conformational shift. It has been shown that this binding occurs independent of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
ADP Ribose
Adenosine diphosphate ribose (ADPR) is an ester molecule formed into chains by the enzyme poly ADP ribose polymerase. ADPR is created from cyclic ADP-ribose (cADPR) by the CD38 enzyme using nicotinamide adenine dinucleotide (NAD+) as a cofactor. ADPR binds to and activates the TRPM2 ion channel. ADPR is the most potent agonist of the TRPM2 channel. cADPR also binds to TPRM2, and the action of both molecules is synergistic, with both molecules enhancing the action of the other molecule in activating the TRPM2 channel. See also * Adenosine diphosphate * ADP-ribosylation * Ribose Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this com ... References Nucleotides Organophosphates NADH dehydrogenase inhibitors Phosphate esters {{biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
PARG
Poly (ADP-ribose) glycohydrolase is an enzyme that in humans is encoded by the ''PARG'' gene. Poly (ADP-ribose) glycohydrolase (PARG) is the major enzyme responsible for the catabolism of poly (ADP-ribose Adenosine diphosphate ribose (ADPR) is an ester molecule formed into chains by the enzyme poly ADP ribose polymerase. ADPR is created from cyclic ADP-ribose (cADPR) by the CD38 enzyme using nicotinamide adenine dinucleotide (NAD+) as a cofactor. ...), a reversible covalent-modifier of chromosomal proteins. The protein is found in many tissues and may be subject to proteolysis generating smaller, active products. References Further reading * * * * * * * * * EC 3.2.1 {{gene-10-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
XRCC1
DNA repair protein XRCC1, also known as X-ray repair cross-complementing protein 1, is a protein that in humans is encoded by the ''XRCC1'' gene. XRCC1 is involved in DNA repair, where it complexes with DNA ligase III. Function XRCC1 is involved in the efficient repair of DNA single-strand breaks formed by exposure to ionizing radiation and alkylating agents. This protein interacts with DNA ligase III, polymerase beta and poly (ADP-ribose) polymerase to participate in the base excision repair pathway. It may play a role in DNA processing during meiogenesis, i.e. during the induction of meiosis and recombination in germ cells. A rare microsatellite polymorphism in this gene is associated with cancer in patients of varying radiosensitivity. The XRCC1 protein does not have enzymatic activity, but acts as a scaffolding protein that interacts with multiple repair enzymes. The scaffolding allows these repair enzymes to then carry out their enzymatic steps in repairing DNA. XRC ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Polymerase Beta
DNA polymerase beta, also known as POLB, is an enzyme present in eukaryotes. In humans, it is encoded by the ''POLB'' gene. Function In eukaryotic cells, DNA polymerase beta (POLB) performs base excision repair (BER) required for DNA maintenance, replication, recombination, and drug resistance. The mitochondrial DNA of mammalian cells is constantly under attack from oxygen radicals released during ATP production. Mammalian cell mitochondria contain an efficient base excision repair system employing POLB that removes some frequent oxidative DNA damages. POLB thus has a key role in maintaining the stability of the mitochondrial genome. An analysis of the fidelity of DNA replication by polymerase beta in the neurons from young and very aged mice indicated that aging has no significant effect on the fidelity of DNA synthesis by polymerase beta. This finding was considered to provide evidence against the error catastrophe theory of aging. Base excision repair Cabelof ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Ligase
DNA ligase is a specific type of enzyme, a ligase, () that facilitates the joining of DNA strands together by catalyzing the formation of a phosphodiester bond. It plays a role in repairing single-strand breaks in duplex DNA in living organisms, but some forms (such as DNA ligase IV) may specifically repair double-strand breaks (i.e. a break in both complementary strands of DNA). Single-strand breaks are repaired by DNA ligase using the complementary strand of the double helix as a template, with DNA ligase creating the final phosphodiester bond to fully repair the DNA. DNA ligase is used in both DNA repair and DNA replication (see '' Mammalian ligases''). In addition, DNA ligase has extensive use in molecular biology laboratories for recombinant DNA experiments (see '' Research applications''). Purified DNA ligase is used in gene cloning to join DNA molecules together to form recombinant DNA. Enzymatic mechanism The mechanism of DNA ligase is to form two covalent p ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Adenosine Diphosphate Ribose
Adenosine diphosphate ribose (ADPR) is an ester molecule formed into chains by the enzyme poly ADP ribose polymerase. ADPR is created from cyclic ADP-ribose (cADPR) by the CD38 enzyme using nicotinamide adenine dinucleotide (NAD+) as a cofactor. ADPR binds to and activates the TRPM2 ion channel. ADPR is the most potent agonist of the TRPM2 channel. cADPR also binds to TPRM2, and the action of both molecules is synergistic, with both molecules enhancing the action of the other molecule in activating the TRPM2 channel. See also * Adenosine diphosphate * ADP-ribosylation * Ribose Ribose is a simple sugar and carbohydrate with molecular formula C5H10O5 and the linear-form composition H−(C=O)−(CHOH)4−H. The naturally-occurring form, , is a component of the ribonucleotides from which RNA is built, and so this com ... References Nucleotides Organophosphates NADH dehydrogenase inhibitors Phosphate esters {{biochemistry-stub ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymeric
A polymer (; Greek '' poly-'', "many" + ''-mer'', "part") is a substance or material consisting of very large molecules called macromolecules, composed of many repeating subunits. Due to their broad spectrum of properties, both synthetic and natural polymers play essential and ubiquitous roles in everyday life. Polymers range from familiar synthetic plastics such as polystyrene to natural biopolymers such as DNA and proteins that are fundamental to biological structure and function. Polymers, both natural and synthetic, are created via polymerization of many small molecules, known as monomers. Their consequently large molecular mass, relative to small molecule compounds, produces unique physical properties including toughness, high elasticity, viscoelasticity, and a tendency to form amorphous and semicrystalline structures rather than crystals. The term "polymer" derives from the Greek word πολύς (''polus'', meaning "many, much") and μέρος (''meros'', meani ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell Nucleus
The cell nucleus (pl. nuclei; from Latin or , meaning ''kernel'' or ''seed'') is a membrane-bound organelle found in eukaryotic cells. Eukaryotic cells usually have a single nucleus, but a few cell types, such as mammalian red blood cells, have no nuclei, and a few others including osteoclasts have many. The main structures making up the nucleus are the nuclear envelope, a double membrane that encloses the entire organelle and isolates its contents from the cellular cytoplasm; and the nuclear matrix, a network within the nucleus that adds mechanical support. The cell nucleus contains nearly all of the cell's genome. Nuclear DNA is often organized into multiple chromosomes – long stands of DNA dotted with various proteins, such as histones, that protect and organize the DNA. The genes within these chromosomes are structured in such a way to promote cell function. The nucleus maintains the integrity of genes and controls the activities of the cell by regulating gene ex ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Bond Cleavage
In chemistry, bond cleavage, or bond fission, is the splitting of chemical bonds. This can be generally referred to as dissociation when a molecule is cleaved into two or more fragments. In general, there are two classifications for bond cleavage: ''homo''lytic and ''hetero''lytic, depending on the nature of the process. The triplet and singlet excitation energies of a sigma bond can be used to determine if a bond will follow the homolytic or heterolytic pathway. A metal−metal sigma bond is an exception because the bond's excitation energy is extremely high, thus cannot be used for observation purposes. In some cases, bond cleavage requires catalysts. Due to the high bond-dissociation energy of C-H bonds, around , a large amount of energy is required to cleave the hydrogen atom from the carbon and bond a different atom to the carbon. Homolytic cleavage In homolytic cleavage, or homolysis, the two electrons in a cleaved covalent bond are divided equally between t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Allosteric Regulation
In biochemistry, allosteric regulation (or allosteric control) is the regulation of an enzyme by binding an effector molecule at a site other than the enzyme's active site. The site to which the effector binds is termed the ''allosteric site'' or ''regulatory site''. Allosteric sites allow effectors to bind to the protein, often resulting in a conformational change and/or a change in protein dynamics. Effectors that enhance the protein's activity are referred to as ''allosteric activators'', whereas those that decrease the protein's activity are called ''allosteric inhibitors''. Allosteric regulations are a natural example of control loops, such as feedback from downstream products or feedforward from upstream substrates. Long-range allostery is especially important in cell signaling. Allosteric regulation is also particularly important in the cell's ability to adjust enzyme activity. The term ''allostery'' comes from the Ancient Greek ''allos'' (), "other", and ''stereos ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Structural Motif
In a chain-like biological molecule, such as a protein or nucleic acid, a structural motif is a common three-dimensional structure that appears in a variety of different, evolutionarily unrelated molecules. A structural motif does not have to be associated with a sequence motif; it can be represented by different and completely unrelated sequences in different proteins or RNA. In nucleic acids Depending upon the sequence and other conditions, nucleic acids can form a variety of structural motifs which is thought to have biological significance. ;Stem-loop: Stem-loop intramolecular base pairing is a pattern that can occur in single-stranded DNA or, more commonly, in RNA. The structure is also known as a hairpin or hairpin loop. It occurs when two regions of the same strand, usually complementary in nucleotide sequence when read in opposite directions, base-pair to form a double helix that ends in an unpaired loop. The resulting structure is a key building block of many ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |