|
Linkage Disequilibrium
Linkage disequilibrium, often abbreviated to LD, is a term in population genetics referring to the association of genes, usually linked genes, in a population. It has become an important tool in medical genetics and other fields In defining LD, it is important first to distinguish the two very different concepts, linkage disequilibrium and linkage (genetic linkage). Linkage disequilibrium refers to the association of genes ''in a population.'' Linkage, on the other hand, tells us whether genes are on the same chromosome ''in an individual''. There is no necessary relationship between the two. Genes that are closely linked may or may not be associated in populations. Looking at parents and offspring, if genes at closely linked loci are together in the parent then they will usually be together in the offspring. But looking at individuals in a population with no known common ancestry, it is much more difficult to see any relationships. To give a concrete, although imaginary, example i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
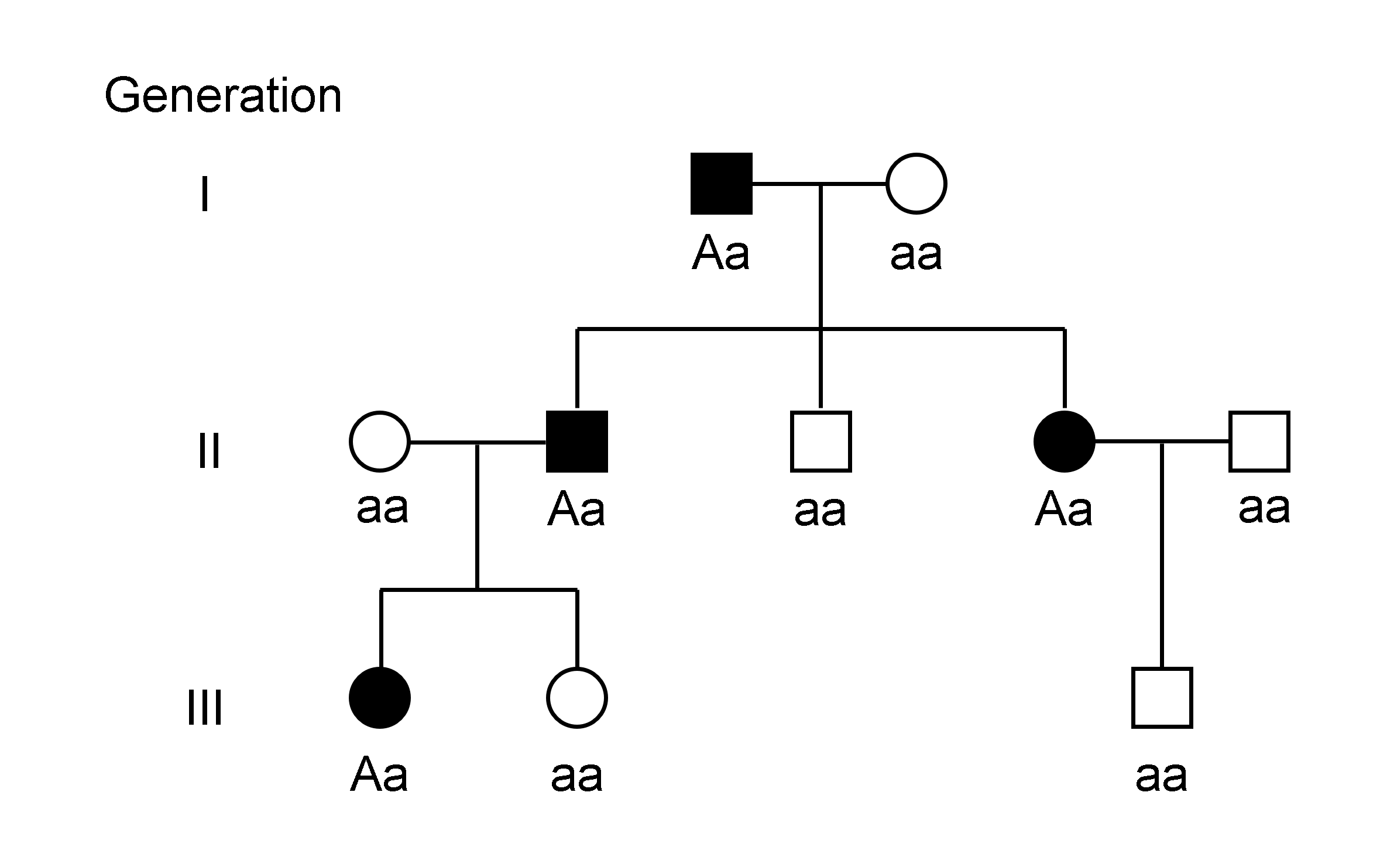
Genetic Linkage
Genetic linkage is the tendency of Nucleic acid sequence, DNA sequences that are close together on a chromosome to be inherited together during the meiosis phase of sexual reproduction. Two Genetic marker, genetic markers that are physically near to each other are unlikely to be separated onto different Chromatid, chromatids during chromosomal crossover, and are therefore said to be more ''linked'' than markers that are far apart. In other words, the nearer two Gene, genes are on a chromosome, the lower the chance of Genetic recombination, recombination between them, and the more likely they are to be inherited together. Markers on different chromosomes are perfectly ''unlinked'', although the penetrance of potentially deleterious alleles may be influenced by the presence of other alleles, and these other alleles may be located on other chromosomes than that on which a particular potentially deleterious allele is located. Genetic linkage is the most prominent exception to Gregor M ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Particulate Inheritance
Particulate inheritance is a pattern of inheritance discovered by Mendelian genetics theorists, such as William Bateson, Ronald Fisher or Gregor Mendel himself, showing that phenotypic traits can be passed from generation to generation through "discrete particles" known as genes, which can keep their ability to be expressed while not always appearing in a descending generation. Scientific developments leading up to the theory Early in the 19th century, scientists had already recognized that Earth has been inhabited by living creatures for a very long time. On the other hand, they did not understand what mechanisms actually drove biological diversity. They also did not understand how physical traits are inherited from one generation to the next. Blending inheritance was the common ideal at the time, but was later discredited by the experiments of Gregor Mendel. Mendel proposed the theory of particulate inheritance by using pea plants (''Pisum sativum'') to explain how variatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
HapMap
The International HapMap Project was an organization that aimed to develop a haplotype map (HapMap) of the human genome, to describe the common patterns of human genetic variation. HapMap is used to find genetic variants affecting health, disease and responses to drugs and environmental factors. The information produced by the project is made freely available for research. The International HapMap Project is a collaboration among researchers at academic centers, non-profit biomedical research groups and private companies in Canada, China (including Hong Kong), Japan, Nigeria, the United Kingdom, and the United States. It officially started with a meeting on October 27 to 29, 2002, and was expected to take about three years. It comprises three phases; the complete data obtained in Phase I were published on 27 October 2005. The analysis of the Phase II dataset was published in October 2007. The Phase III dataset was released in spring 2009 and the publication presenting the final ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
1000 Genomes Project
The 1000 Genomes Project (1KGP), taken place from January 2008 to 2015, was an international research effort to establish the most detailed catalogue of human genetic variation at the time. Scientists planned to sequence the genomes of at least one thousand anonymous healthy participants from a number of different ethnic groups within the following three years, using advancements in newly developed technologies. In 2010, the project finished its pilot phase, which was described in detail in a publication in the journal ''Nature''. In 2012, the sequencing of 1092 genomes was announced in a ''Nature'' publication. In 2015, two papers in ''Nature'' reported results and the completion of the project and opportunities for future research. Many rare variations, restricted to closely related groups, were identified, and eight structural-variation classes were analyzed. The project united multidisciplinary research teams from institutes around the world, including China, Italy, Japan, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Haploview
Haploview is a commonly used bioinformatics software which is designed to analyze and visualize patterns of linkage disequilibrium (LD) in genetic data. Haploview can also perform association studies, choosing tag SNPs{{cite journal, author1=de Bakker P. I. , author2=Yelensky R. , author3=Pe'er I. , author4=Gabriel S. B. , author5=Daly M. J. , author6=Altshuler D. , title=Efficiency and power in genetic association studies, journal=Nature Genetics, year=2005, volume=37, pages=1217–23, doi=10.1038/ng1669, pmid=16244653, issue=11 and estimating haplotype frequencies. Haploview is developed and maintained by Dr. Mark Daly's lab at the MIT/Harvard Broad Institute. Haploview currently supports the following functionalities: * LD & haplotype block analysis * Haplotype population frequency estimation * Single SNP and haplotype association tests * Permutation testing for association significance * Implementation of Paul de Bakker's Tagger tag SNP selection algorithm * Automatic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DbSNP
The Single Nucleotide Polymorphism Database (dbSNP) is a free public archive for genetic variation within and across different species developed and hosted by the National Center for Biotechnology Information (NCBI) in collaboration with the National Human Genome Research Institute (NHGRI). Although the name of the database implies a collection of one class of polymorphisms only (i.e., single nucleotide polymorphisms (SNPs)), it in fact contains a range of molecular variation: (1) SNPs, (2) short deletion and insertion polymorphisms ( indels/DIPs), (3) microsatellite markers or short tandem repeats (STRs), (4) multinucleotide polymorphisms (MNPs), (5) heterozygous sequences, and (6) named variants. The dbSNP accepts apparently neutral polymorphisms, polymorphisms corresponding to known phenotypes, and regions of no variation. It was created in September 1998 to supplement GenBank, NCBI’s collection of publicly available nucleic acid and protein sequences. In 2017, NCBI stopped ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Ensembl
Ensembl genome database project is a scientific project at the European Bioinformatics Institute, which provides a centralized resource for geneticists, molecular biologists and other researchers studying the genomes of our own species and other vertebrates and model organisms. Ensembl is one of several well known genome browsers for the retrieval of genomic information. Similar databases and browsers are found at National Center for Biotechnology Information, NCBI and UCSC Genome Browser, the University of California, Santa Cruz (UCSC). History The human genome consists of three billion base pairs, which code for approximately 20,000–25,000 genes. However the genome alone is of little use, unless the locations and relationships of individual genes can be identified. One option is manual annotation, whereby a team of scientists tries to locate genes using experimental data from scientific journals and public databases. However this is a slow, painstaking task. The alternative, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
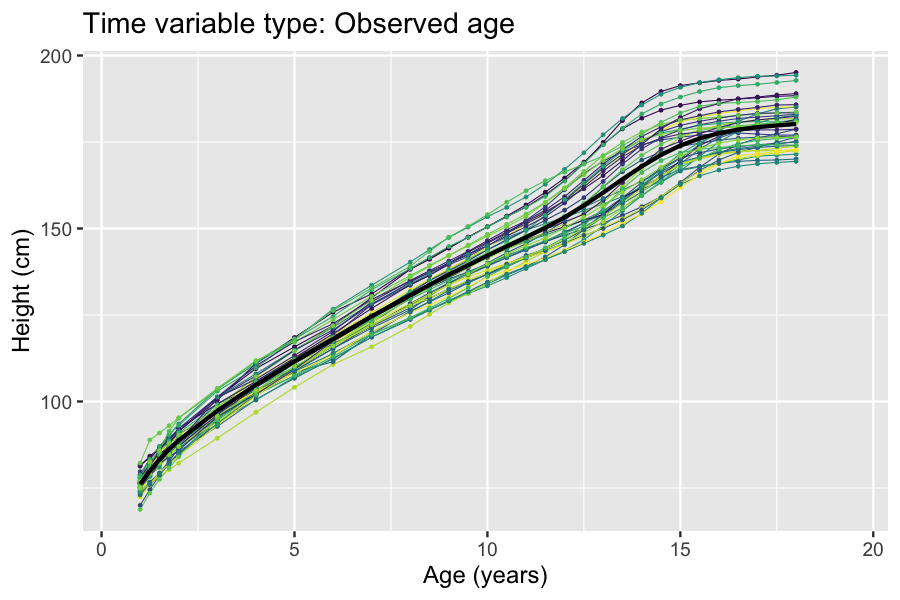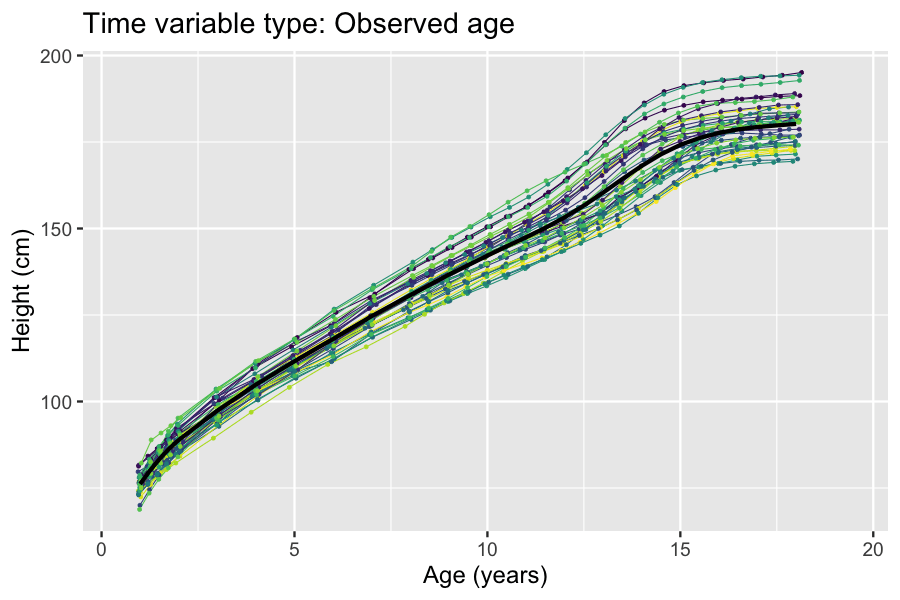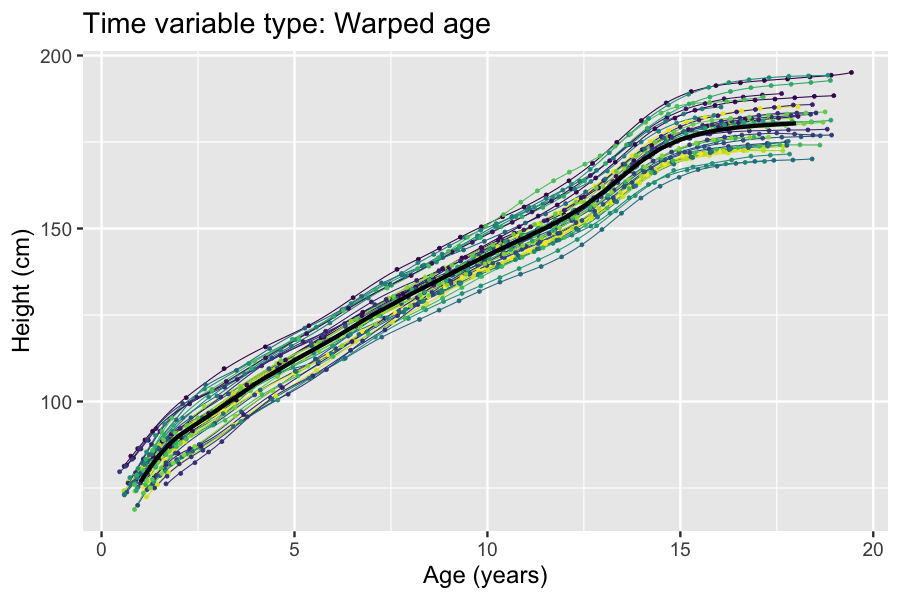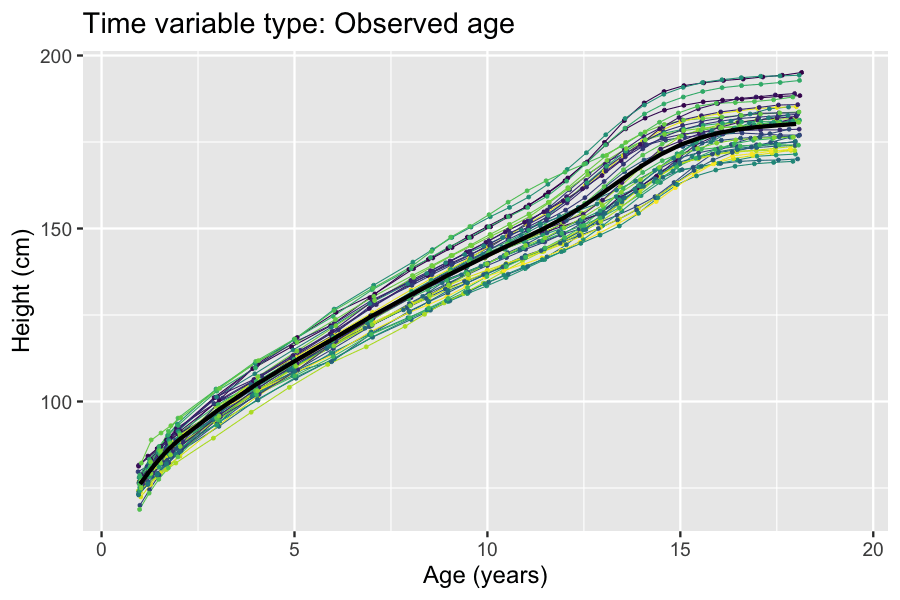
Latent And Observable Variables
In statistics, latent variables (from Latin: present participle of ) are variables that can only be inferred indirectly through a mathematical model from other observable variables that can be directly observed or measured. Such ''latent variable models'' are used in many disciplines, including engineering, medicine, ecology, physics, machine learning/artificial intelligence, natural language processing, bioinformatics, chemometrics, demography, economics, management, political science, psychology and the social sciences. Latent variables may correspond to aspects of physical reality. These could in principle be measured, but may not be for practical reasons. Among the earliest expressions of this idea is Francis Bacon's polemic the ''Novum Organum'', itself a challenge to the more traditional logic expressed in Aristotle's Organon: In this situation, the term ''hidden variables'' is commonly used, reflecting the fact that the variables are meaningful, but not observable. Othe ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as Zygosity, homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plan ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Linkage Disequilibrium Heatmap
Linkage may refer to: * ''Linkage'' (album), by J-pop singer Mami Kawada, released in 2010 *Linkage (graph theory), the maximum min-degree of any of its subgraphs *Linkage (horse), an American Thoroughbred racehorse * Linkage (hierarchical clustering), The linkage criterion determines the distance between sets of observations as a function of the pairwise distances between observations *Linkage (linguistics), a set of languages descended from a former dialect continuum *Linkage (mechanical), assemblies of links designed to manage forces and movement *Linkage (policy), a Cold War policy of the United States of America towards the Soviet Union and Communist China *Linkage (software), a concept in computer programming *Genetic linkage, the tendency of certain genes to be inherited together *Glycosidic linkage, a type of covalent bond that joins a carbohydrate (sugar) molecule to another chemical group *Flux linkage, the total flux passing through a surface formed by a closed conducting ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Heatmap
A heat map (or heatmap) is a 2-dimensional data visualization technique that represents the magnitude of individual values within a dataset as a color. The variation in color may be by hue or brightness, intensity. In some applications such as crime analytics or website click-tracking, color is used to represent the ''density'' of data points rather than a value associated with each point. "Heat map" is a relatively new term, but the practice of shading matrices has existed for over a century. History Heat maps originated in 2D displays of the values in a data matrix. Larger values were represented by small dark gray or black squares (pixels) and smaller values by lighter squares. The earliest known example dates to 1873, when Toussaint Loua used a hand-drawn and colored shaded matrix to visualize social statistics across the districts of Paris. The idea of reordering rows and columns to reveal structure in a data matrix, known as Seriation (statistics), seriation, was introduc ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetic Drift
Genetic drift, also known as random genetic drift, allelic drift or the Wright effect, is the change in the Allele frequency, frequency of an existing gene variant (allele) in a population due to random chance. Genetic drift may cause gene variants to disappear completely and thereby reduce genetic variation. It can also cause initially rare alleles to become much more frequent and even fixed. When few copies of an allele exist, the effect of genetic drift is more notable, and when many copies exist, the effect is less notable (due to the law of large numbers). In the middle of the 20th century, vigorous debates occurred over the relative importance of natural selection versus neutral processes, including genetic drift. Ronald Fisher, who explained natural selection using Mendelian inheritance, Mendelian genetics, held the view that genetic drift plays at most a minor role in evolution, and this remained the dominant view for several decades. In 1968, population geneticist Mot ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |