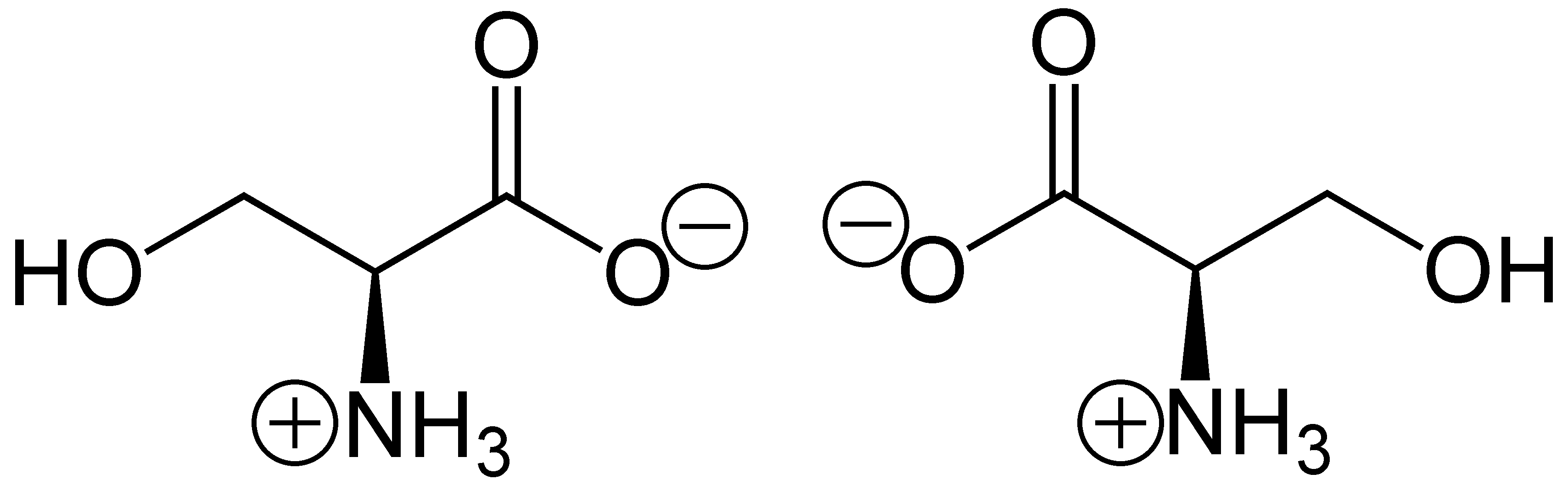|
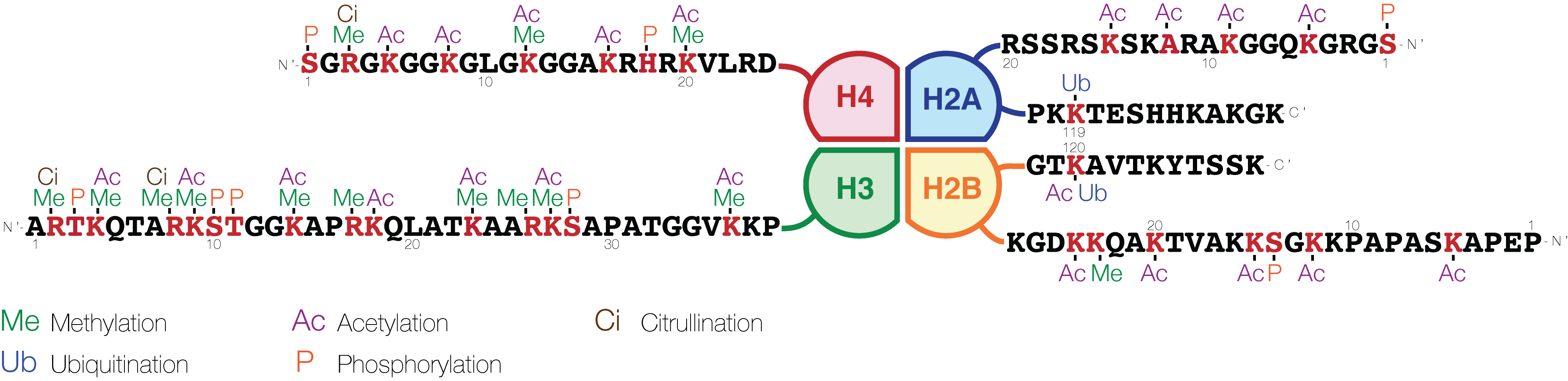
Histone Code
The histone code is a hypothesis that the transcription of genetic information encoded in DNA is in part regulated by chemical modifications (known as ''histone marks'') to histone proteins, primarily on their unstructured ends. Together with similar modifications such as DNA methylation it is part of the epigenetic code. Histones associate with DNA to form nucleosomes, which themselves bundle to form chromatin fibers, which in turn make up the more familiar chromosome. Histones are globular proteins with a flexible N-terminus (taken to be the tail) that protrudes from the nucleosome. Many of the histone tail modifications correlate very well to chromatin structure and both histone modification state and chromatin structure correlate well to gene expression levels. The critical concept of the histone code hypothesis is that the histone modifications serve to recruit other proteins by specific recognition of the modified histone via protein domains specialized for such purpose ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hypothesis
A hypothesis (plural hypotheses) is a proposed explanation for a phenomenon. For a hypothesis to be a scientific hypothesis, the scientific method requires that one can test it. Scientists generally base scientific hypotheses on previous observations that cannot satisfactorily be explained with the available scientific theories. Even though the words "hypothesis" and "theory" are often used interchangeably, a scientific hypothesis is not the same as a scientific theory. A working hypothesis is a provisionally accepted hypothesis proposed for further research in a process beginning with an educated guess or thought. A different meaning of the term ''hypothesis'' is used in formal logic, to denote the antecedent of a proposition; thus in the proposition "If ''P'', then ''Q''", ''P'' denotes the hypothesis (or antecedent); ''Q'' can be called a consequent. ''P'' is the assumption in a (possibly counterfactual) ''What If'' question. The adjective ''hypothetical'', meaning ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Serine
Serine (symbol Ser or S) is an α-amino acid that is used in the biosynthesis of proteins. It contains an α- amino group (which is in the protonated − form under biological conditions), a carboxyl group (which is in the deprotonated − form under biological conditions), and a side chain consisting of a hydroxymethyl group, classifying it as a polar amino acid. It can be synthesized in the human body under normal physiological circumstances, making it a nonessential amino acid. It is encoded by the codons UCU, UCC, UCA, UCG, AGU and AGC. Occurrence This compound is one of the naturally occurring proteinogenic amino acids. Only the L- stereoisomer appears naturally in proteins. It is not essential to the human diet, since it is synthesized in the body from other metabolites, including glycine. Serine was first obtained from silk protein, a particularly rich source, in 1865 by Emil Cramer. Its name is derived from the Latin for silk, ''sericum''. Serine's structure w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Acetyltransferase
Histone acetyltransferases (HATs) are enzymes that acetylate conserved lysine amino acids on histone proteins by transferring an acetyl group from acetyl-CoA to form ε-''N''-acetyllysine. DNA is wrapped around histones, and, by transferring an acetyl group to the histones, genes can be turned on and off. In general, histone acetylation increases gene expression. In general, histone acetylation is linked to transcriptional activation and associated with euchromatin. Euchromatin, which is less densely compact, allows transcription factors to bind more easily to regulatory sites on DNA, causing transcriptional activation. When it was first discovered, it was thought that acetylation of lysine neutralizes the positive charge normally present, thus reducing affinity between histone and (negatively charged) DNA, which renders DNA more accessible to transcription factors. Research has emerged, since, to show that lysine acetylation and other posttranslational modifications of his ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Acetylation
: In organic chemistry, acetylation is an organic esterification reaction with acetic acid. It introduces an acetyl group into a chemical compound. Such compounds are termed ''acetate esters'' or simply '' acetates''. Deacetylation is the opposite reaction, the removal of an acetyl group from a chemical compound. Organic synthesis Acetate esters and acetamides are generally prepared by acetylations. Acetylations are often used in making C-acetyl bonds in Friedel-Crafts reactions. Carbanions and their equivalents are susceptible to acetylations. Acetylation reagents Many acetylations are achieved using these three reagents: *Acetic anhydride. This reagent is common in the laboratory; its use cogenerates acetic acid. * Acetyl chloride. This reagent is also common in the laboratory, but its use cogenerates hydrogen chloride, which can be undesirable. * Ketene. At one time acetic anhydride was prepared by the reaction of ketene with acetic acid: :H2C=C=O + CH3COOH -> (CH3CO)2 ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Non-homologous End Joining
Non-homologous end joining (NHEJ) is a pathway that repairs double-strand breaks in DNA. NHEJ is referred to as "non-homologous" because the break ends are directly ligated without the need for a homologous template, in contrast to homology directed repair(HDR), which requires a homologous sequence to guide repair. NHEJ is active in both non-dividing and proliferating cells, while HDR is not readily accessible in non-dividing cells. The term "non-homologous end joining" was coined in 1996 by Moore and Haber. NHEJ is typically guided by short homologous DNA sequences called microhomologies. These microhomologies are often present in single-stranded overhangs on the ends of double-strand breaks. When the overhangs are perfectly compatible, NHEJ usually repairs the break accurately. Imprecise repair leading to loss of nucleotides can also occur, but is much more common when the overhangs are not compatible. Inappropriate NHEJ can lead to translocations and telomere fusion, hallma ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Damage (naturally Occurring)
DNA damage is an alteration in the chemical structure of DNA, such as a break in a strand of DNA, a nucleobase missing from the backbone of DNA, or a chemically changed base such as 8-OHdG. DNA damage can occur naturally or via environmental factors, but is distinctly different from mutation, although both are types of error in DNA. DNA damage is an abnormal chemical structure in DNA, while a mutation is a change in the sequence of base pairs. DNA damages cause changes in the structure of the genetic material and prevents the replication mechanism from functioning and performing properly. The DNA damage response (DDR) is a complex signal transduction pathway which recognizes when DNA is damaged and initiates the cellular response to the damage. DNA damage and mutation have different biological consequences. While most DNA damages can undergo DNA repair, such repair is not 100% efficient. Un-repaired DNA damages accumulate in non-replicating cells, such as cells in the brains o ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Homologous Recombination
Homologous recombination is a type of genetic recombination in which genetic information is exchanged between two similar or identical molecules of double-stranded or single-stranded nucleic acids (usually DNA as in cellular organisms but may be also RNA in viruses). Homologous recombination is widely used by cells to accurately DNA repair harmful breaks that occur on both strands of DNA, known as double-strand breaks (DSB), in a process called homologous recombinational repair (HRR). Homologous recombination also produces new combinations of DNA sequences during meiosis, the process by which eukaryotes make gamete cells, like sperm and egg cells in animals. These new combinations of DNA represent genetic variation in offspring, which in turn enables populations to adapt during the course of evolution. Homologous recombination is also used in horizontal gene transfer to exchange genetic material between different strains and species of bacteria and viruses. Horizon ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K36me
H3K36me is an epigenetic modification to the DNA packaging protein Histone H3, specifically, the mono-methylation at the 36th lysine residue of the histone H3 protein. There are diverse modifications at H3K36, such as phosphorylation, methylation, acetylation, and ubiquitylation, which have many important biological processes. The methylation of H3K36 has particularly had effects in transcriptional repression, alternative splicing, dosage compensation, DNA replication and repair, DNA methylation, and the transmission of the memory of gene expression from parents to offspring during development. Nomenclature H3K36me2 indicates dimethylation of lysine 36 on histone H3 protein subunit: Lysine methylation This diagram shows the progressive methylation of a lysine residue. The mono-methylation denotes the methylation present in H3K36me1. Lysine methylation is the addition of a methyl group to the lysine of histone proteins. This occurs via histone lysine methyltransferase (HMT ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
DNA Repair
DNA repair is a collection of processes by which a cell identifies and corrects damage to the DNA molecules that encode its genome. In human cells, both normal metabolic activities and environmental factors such as radiation can cause DNA damage, resulting in tens of thousands of individual molecular lesions per cell per day. Many of these lesions cause structural damage to the DNA molecule and can alter or eliminate the cell's ability to transcribe the gene that the affected DNA encodes. Other lesions induce potentially harmful mutations in the cell's genome, which affect the survival of its daughter cells after it undergoes mitosis. As a consequence, the DNA repair process is constantly active as it responds to damage in the DNA structure. When normal repair processes fail, and when cellular apoptosis does not occur, irreparable DNA damage may occur, including double-strand breaks and DNA crosslinkages (interstrand crosslinks or ICLs). This can eventually lead to malig ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
H3K9me3
H3K9me3 is an epigenetic modification to the DNA packaging protein Histone H3. It is a mark that indicates the tri-methylation at the 9th lysine residue of the histone H3 protein and is often associated with heterochromatin. Nomenclature H3K9me3 indicates trimethylation of lysine 9 on histone H3 protein subunit: Lysine Methylation This diagram shows the progressive methylation of a lysine residue. The tri-methylation denotes the methylation present in H3K9me3 . Understanding histone modifications The genomic DNA of eukaryotic cells is wrapped around special protein molecules known as Histones. The complexes formed by the looping of the DNA are known as chromatin. The basic structural unit of chromatin is the nucleosome: this consists of the core octamer of histones (H2A, H2B, H3 and H4) as well as a linker histone and about 180 base pairs of DNA. These core histones are rich in lysine and arginine residues. The carboxyl (C) terminal end of these histones contribute to hi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Methylation
In the chemical sciences, methylation denotes the addition of a methyl group on a substrate, or the substitution of an atom (or group) by a methyl group. Methylation is a form of alkylation, with a methyl group replacing a hydrogen atom. These terms are commonly used in chemistry, biochemistry, soil science, and the biological sciences. In biological systems, methylation is catalyzed by enzymes; such methylation can be involved in modification of heavy metals, regulation of gene expression, regulation of protein function, and RNA processing. In vitro methylation of tissue samples is also one method for reducing certain histological staining artifacts. The reverse of methylation is demethylation. In biology In biological systems, methylation is accomplished by enzymes. Methylation can modify heavy metals, regulate gene expression, RNA processing and protein function. It has been recognized as a key process underlying epigenetics. Methanogenesis Methanogenesis, the proces ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Histone Modifications
In biology, histones are highly basic proteins abundant in lysine and arginine residues that are found in eukaryotic cell nuclei. They act as spools around which DNA winds to create structural units called nucleosomes. Nucleosomes in turn are wrapped into 30-nanometer fibers that form tightly packed chromatin. Histones prevent DNA from becoming tangled and protect it from DNA damage. In addition, histones play important roles in gene regulation and DNA replication. Without histones, unwound DNA in chromosomes would be very long. For example, each human cell has about 1.8 meters of DNA if completely stretched out; however, when wound about histones, this length is reduced to about 90 micrometers (0.09 mm) of 30 nm diameter chromatin fibers. There are five families of histones which are designated H1/H5 (linker histones), H2, H3, and H4 (core histones). The nucleosome core is formed of two H2A-H2B dimers and a H3-H4 tetramer. The tight wrapping of DNA around histones ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |