|
Cycling Probe Technology
Cycling probe technology (CPT) is a molecular biological technique for detecting specific DNA sequences. CPT operates under isothermal conditions. In some applications, CPT offers an alternative to PCR. However, unlike PCR, CPT does not generate multiple copies of the target DNA itself, and the amplification of the signal is linear, in contrast to the exponential amplification of the target DNA in PCR. CPT uses a sequence specific chimeric probe which hybridizes to a complementary target DNA sequence and becomes a substrate for RNase H. Cleavage occurs at the RNA internucleotide linkages and results in dissociation of the probe from the target, thereby making it available for the next probe molecule. Integrated electrokinetic systems have been developed for use in CPT. Probe Cycling probe technology makes use of a chimeric nucleic acid probe to detect the presence of a particular DNA sequence. The chimeric probe consists of an RNA segment sandwiched between two DNA segments ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Polymerase Chain Reaction Optimization
The polymerase chain reaction (PCR) is a commonly used molecular biology tool for amplifying DNA, and various techniques for PCR optimization which have been developed by molecular biologists to improve PCR performance and minimize failure. Contamination and PCR The PCR method is extremely sensitive, requiring only a few DNA molecules in a single reaction for amplification across several orders of magnitude. Therefore, adequate measures to avoid contamination from any DNA present in the lab environment (bacteria, viruses, or human sources) are required. Because products from previous PCR amplifications are a common source of contamination, many molecular biology labs have implemented procedures that involve dividing the lab into separate areas. One lab area is dedicated to preparation and handling of pre-PCR reagents and the setup of the PCR reaction, and another area to post-PCR processing, such as gel electrophoresis or PCR product purification. For the setup of PCR reactions, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is the branch of biology that seeks to understand the molecular basis of biological activity in and between cells, including biomolecular synthesis, modification, mechanisms, and interactions. The study of chemical and physical structure of biological macromolecules is known as molecular biology. Molecular biology was first described as an approach focused on the underpinnings of biological phenomena - uncovering the structures of biological molecules as well as their interactions, and how these interactions explain observations of classical biology. In 1945 the term molecular biology was used by physicist William Astbury. In 1953 Francis Crick, James Watson, Rosalind Franklin, and colleagues, working at Medical Research Council unit, Cavendish laboratory, Cambridge (now the MRC Laboratory of Molecular Biology), made a double helix model of DNA which changed the entire research scenario. They proposed the DNA structure based on previous research done by R ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
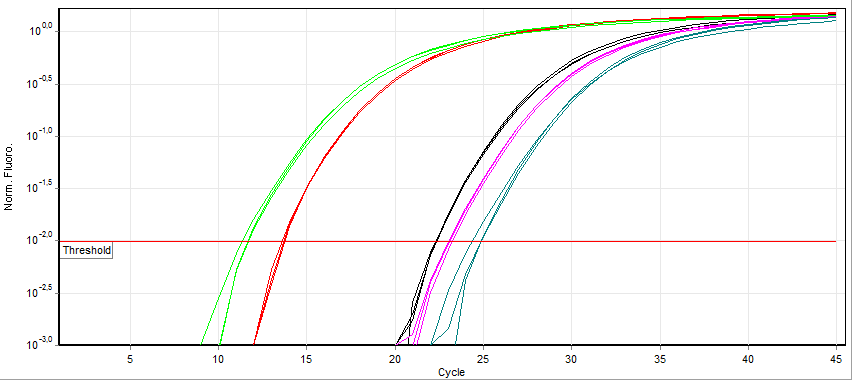
Real-time Polymerase Chain Reaction
A real-time polymerase chain reaction (real-time PCR, or qPCR) is a laboratory technique of molecular biology based on the polymerase chain reaction (PCR). It monitors the amplification of a targeted DNA molecule during the PCR (i.e., in real time), not at its end, as in conventional PCR. Real-time PCR can be used quantitatively (quantitative real-time PCR) and semi-quantitatively (i.e., above/below a certain amount of DNA molecules) (semi-quantitative real-time PCR). Two common methods for the detection of PCR products in real-time PCR are (1) non-specific fluorescent dyes that Intercalation (biochemistry), intercalate with any double-stranded DNA and (2) sequence-specific DNA probes consisting of oligonucleotides that are labelled with a fluorescence, fluorescent reporter, which permits detection only after nucleic acid hybridisation, hybridization of the probe with its complementary sequence. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Beacon
Molecular beacons, or molecular beacon probes, are oligonucleotide hybridization probes that can report the presence of specific nucleic acids in homogenous solutions. Molecular beacons are hairpin-shaped molecules with an internally quenched fluorophore whose fluorescence is restored when they bind to a target nucleic acid sequence. This is a novel non-radioactive method for detecting specific sequences of nucleic acids. They are useful in situations where it is either not possible or desirable to isolate the probe-target hybrids from an excess of the hybridization probes. Molecular beacon probes A typical molecular beacon probe is 25 nucleotides long. The middle 15 nucleotides are complementary to the target DNA or RNA and do not base pair with one another, while the five nucleotides at each terminus are complementary to each other rather than to the target DNA. A typical molecular beacon structure can be divided in 4 parts: 1) loop, an 18–30 base pair region of the molecula ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Rate Equation
In chemistry, the rate law or rate equation for a reaction is an equation that links the initial or forward reaction rate with the concentrations or pressures of the reactants and constant parameters (normally rate coefficients and partial reaction orders). For many reactions, the initial rate is given by a power law such as :v_0\; =\; k mathrmx mathrmy where and express the concentration of the species and usually in moles per liter (molarity, ). The exponents and are the partial ''orders of reaction'' for and and the ''overall'' reaction order is the sum of the exponents. These are often positive integers, but they may also be zero, fractional, or negative. The constant is the reaction rate constant or ''rate coefficient'' of the reaction. Its value may depend on conditions such as temperature, ionic strength, surface area of an adsorbent, or light irradiation. If the reaction goes to completion, the rate equation for the reaction rate v\; =\; k cex cey applies thr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Antimicrobial Resistance
Antimicrobial resistance (AMR) occurs when microbes evolve mechanisms that protect them from the effects of antimicrobials. All classes of microbes can evolve resistance. Fungi evolve antifungal resistance. Viruses evolve antiviral resistance. Protozoa evolve antiprotozoal resistance, and bacteria evolve antibiotic resistance. Those bacteria that are considered extensively drug resistant (XDR) or totally drug-resistant (TDR) are sometimes called "superbugs".A.-P. Magiorakos, A. Srinivasan, R. B. Carey, Y. Carmeli, M. E. Falagas, C. G. Giske, S. Harbarth, J. F. Hinndler ''et al''Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria... Clinical Microbiology and Infection, Vol 8, Iss. 3 first published 27 July 2011 ia Wiley Online Library Retrieved 28 August 2020 Although antimicrobial resistance is a naturally-occurring process, it is often the result of improper usage of the drugs and management of the infections. Antibiotic resistance is a major subset of ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Genetically Modified Organism
A genetically modified organism (GMO) is any organism whose genetic material has been altered using genetic engineering techniques. The exact definition of a genetically modified organism and what constitutes genetic engineering varies, with the most common being an organism altered in a way that "does not occur naturally by mating and/or natural recombination". A wide variety of organisms have been genetically modified (GM), from animals to plants and microorganisms. Genes have been transferred within the same species, across species (creating transgenic organisms), and even across kingdoms. New genes can be introduced, or endogenous genes can be enhanced, altered, or knocked out. Creating a genetically modified organism is a multi-step process. Genetic engineers must isolate the gene they wish to insert into the host organism and combine it with other genetic elements, including a promoter and terminator region and often a selectable marker. A number of techniques ar ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
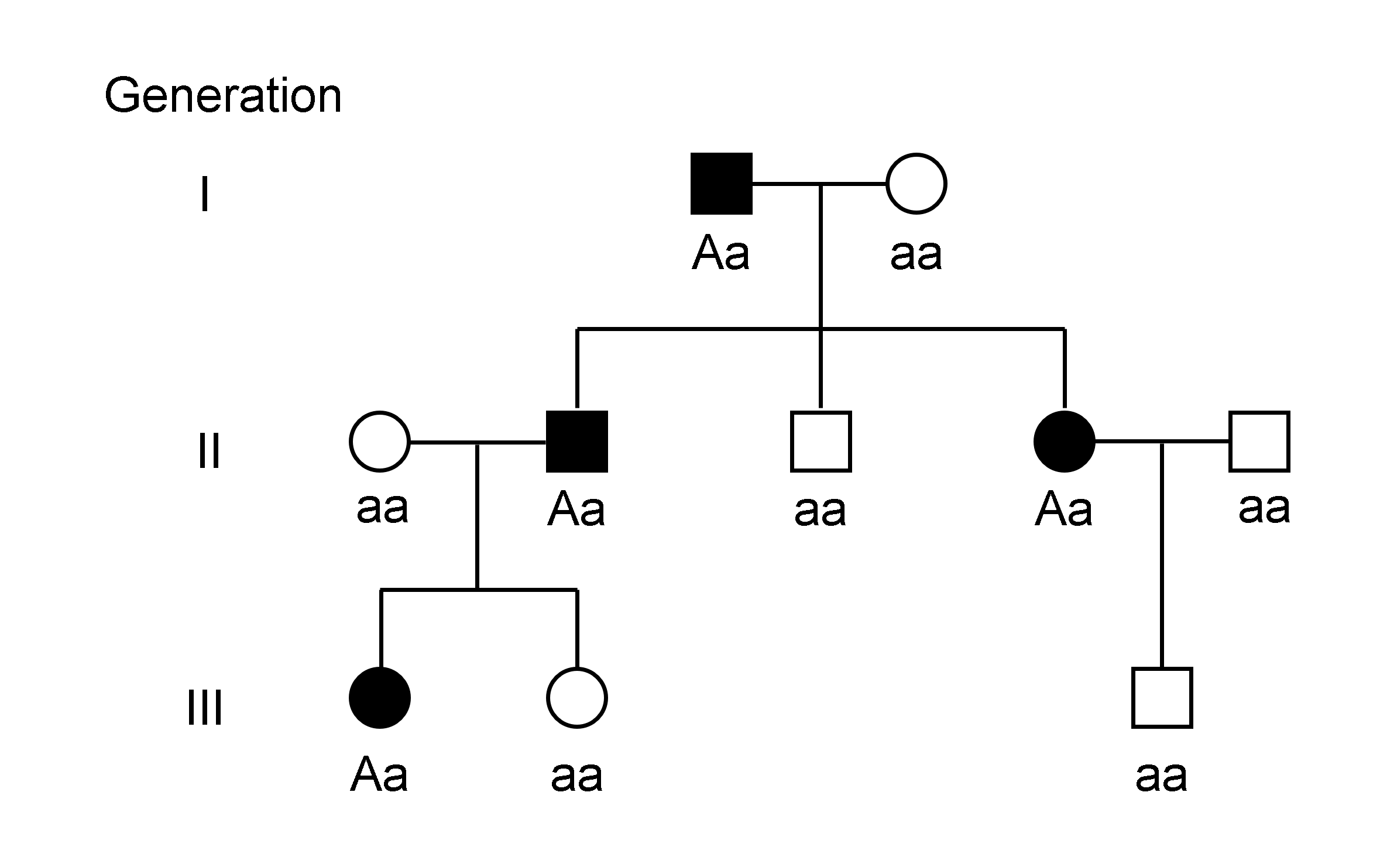
Genotype
The genotype of an organism is its complete set of genetic material. Genotype can also be used to refer to the alleles or variants an individual carries in a particular gene or genetic location. The number of alleles an individual can have in a specific gene depends on the number of copies of each chromosome found in that species, also referred to as ploidy. In diploid species like humans, two full sets of chromosomes are present, meaning each individual has two alleles for any given gene. If both alleles are the same, the genotype is referred to as homozygous. If the alleles are different, the genotype is referred to as heterozygous. Genotype contributes to phenotype, the observable traits and characteristics in an individual or organism. The degree to which genotype affects phenotype depends on the trait. For example, the petal color in a pea plant is exclusively determined by genotype. The petals can be purple or white depending on the alleles present in the pea plant. Howev ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
RNAse
Ribonuclease (commonly abbreviated RNase) is a type of nuclease that catalyzes the degradation of RNA into smaller components. Ribonucleases can be divided into endoribonucleases and exoribonucleases, and comprise several sub-classes within the EC 2.7 (for the phosphorolytic enzymes) and 3.1 (for the hydrolytic enzymes) classes of enzymes. Function All organisms studied contain many RNases of two different classes, showing that RNA degradation is a very ancient and important process. As well as clearing of cellular RNA that is no longer required, RNases play key roles in the maturation of all RNA molecules, both messenger RNAs that carry genetic material for making proteins and non-coding RNAs that function in varied cellular processes. In addition, active RNA degradation systems are the first defense against RNA viruses and provide the underlying machinery for more advanced cellular immune strategies such as RNAi. Some cells also secrete copious quantities of non-specific ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Gel Electrophoresis
Gel electrophoresis is a method for separation and analysis of biomacromolecules ( DNA, RNA, proteins, etc.) and their fragments, based on their size and charge. It is used in clinical chemistry to separate proteins by charge or size (IEF agarose, essentially size independent) and in biochemistry and molecular biology to separate a mixed population of DNA and RNA fragments by length, to estimate the size of DNA and RNA fragments or to separate proteins by charge. Nucleic acid molecules are separated by applying an electric field to move the negatively charged molecules through a matrix of agarose or other substances. Shorter molecules move faster and migrate farther than longer ones because shorter molecules migrate more easily through the pores of the gel. This phenomenon is called sieving. Proteins are separated by the charge in agarose because the pores of the gel are too small to sieve proteins. Gel electrophoresis can also be used for the separation of nanoparticl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Thermal Cycler
The thermal cycler (also known as a thermocycler, PCR machine or DNA amplifier) is a laboratory apparatus most commonly used to amplify segments of DNA via the polymerase chain reaction (PCR). Thermal cyclers may also be used in laboratories to facilitate other temperature-sensitive reactions, including restriction enzyme digestion or rapid diagnostics. The device has a ''thermal block'' with holes where tubes holding the reaction mixtures can be inserted. The cycler then raises and lowers the temperature of the block in discrete, pre-programmed steps. History The earliest thermal cyclers were designed for use with the Klenow fragment of DNA polymerase I. Since this enzyme is destroyed during each heating step of the amplification process, new enzyme had to be added every cycle. This led to a cumbersome machine based on an automated pipettor, with open reaction tubes. Later, the PCR process was adapted to the use of thermostable DNA polymerase from '' Thermus aquaticus'', ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |