|
Chimeric Nuclease
Chimeric nucleases are an example of engineered proteins which must comprise a DNA-binding domain to give sequence specificity and a nuclease domain for DNA cleavage. DNA-binding domains DNA-binding domains including the basic helix-loop-helix, zinc finger, helix-turn-helix and leucine zipper motifs have been used in construction of sequence-specific nucleases. Of these, zinc fingers have been suggested the most important due to their modularity, allowing construction of a tailor-made DNA-binding domain. Nuclease domain The nuclease domain is responsible for physical cleavage of DNA strands and may introduce either single stranded or double-stranded breaks. FokI is an example of a sequence-specific endonuclease whose non-specific nuclease domain introduces double stranded breaks and has been used in a variety of experiments including identification of high- and low-affinity binding sites of transcription factors ''in vitro'', to study recruitment of factors to promoter si ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Engineering
Protein engineering is the process of developing useful or valuable proteins through the design and production of unnatural polypeptides, often by altering amino acid sequences found in nature. It is a young discipline, with much research taking place into the understanding of protein folding and recognition for protein design principles. It has been used to improve the function of many enzymes for industrial catalysis. It is also a product and services market, with an estimated value of $168 billion by 2017. There are two general strategies for protein engineering: rational protein design and directed evolution. These methods are not mutually exclusive; researchers will often apply both. In the future, more detailed knowledge of protein structure and function, and advances in high-throughput screening, may greatly expand the abilities of protein engineering. Eventually, even unnatural amino acids may be included, via newer methods, such as expanded genetic code, that allow e ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
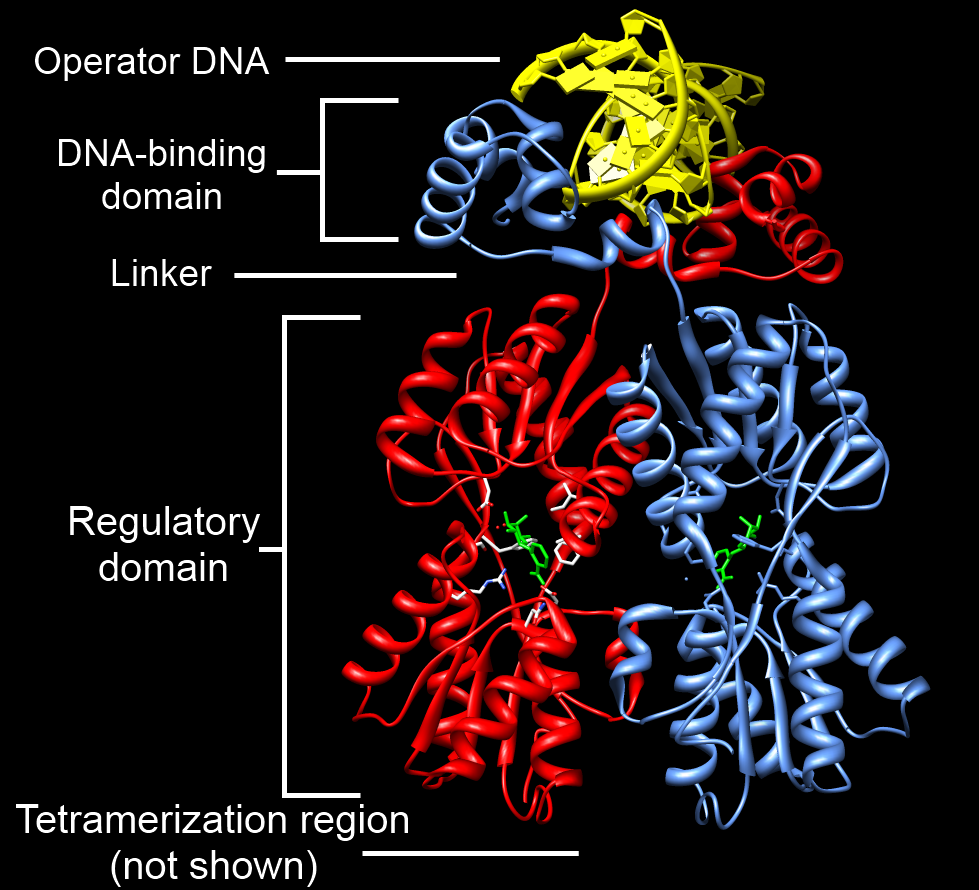
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nuclease
In biochemistry, a nuclease (also archaically known as nucleodepolymerase or polynucleotidase) is an enzyme capable of cleaving the phosphodiester bonds that link nucleotides together to form nucleic acids. Nucleases variously affect single and double stranded breaks in their target molecules. In living organisms, they are essential machinery for many aspects of DNA repair. Defects in certain nucleases can cause genetic instability or immunodeficiency. Nucleases are also extensively used in molecular cloning. There are two primary classifications based on the locus of activity. Exonucleases digest nucleic acids from the ends. Endonucleases act on regions in the ''middle'' of target molecules. They are further subcategorized as deoxyribonucleases and ribonucleases. The former acts on DNA, the latter on RNA. History In the late 1960s, scientists Stuart Linn and Werner Arber isolated examples of the two types of enzymes responsible for phage growth restriction in Escherichi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Basic Helix-loop-helix
Basic or BASIC may refer to: Science and technology * BASIC, a computer programming language * Basic (chemistry), having the properties of a base * Basic access authentication, in HTTP Entertainment * ''Basic'' (film), a 2003 film * Basic, one of the languages in ''Star Wars'' Music * ''Basic'' (Glen Campbell album), 1978 * ''Basic'' (Robert Quine and Fred Maher album), 1984 * ''B.A.S.I.C.'' (Alpinestars album), 2000 * ''Basic'' (Brown Eyed Girls album), 2015 * ''B.A.S.I.C.'' (The Basics album), 2019 Places * Basic, Mississippi, a community in the US * BASIC countries, Brazil, South Africa, India and China in climate change negotiations Organizations * BASIC Bank Limited, government owned bank in Bangladesh * Basic Books, an American publisher Other uses * Basic (cigarette), a brand of cigarettes manufactured by the Altria Group (Philip Morris Company) * Basic (dance move), the dance move that defines the character of a particular dance * Basic (slang), a pejorative t ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) which stabilizes the fold. The term ''zinc finger'' was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. '' Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Helix-turn-helix
Helix-turn-helix is a DNA-binding domain (DBD). The helix-turn-helix (HTH) is a major structural motif capable of binding DNA. Each monomer incorporates two alpha helix, α helices, joined by a short strand of amino acids, that bind to the major groove of DNA. The HTH motif occurs in many proteins that regulate gene expression. It should not be confused with the helix–loop–helix motif. Discovery The discovery of the helix-turn-helix motif was based on similarities between several genes encoding Transcription (genetics), transcription regulatory proteins from bacteriophage lambda and ''Escherichia coli'': Cro, Catabolite activator protein, CAP, and cI protein, λ repressor, which were found to share a common 20–25 amino acid sequence that facilitates DNA recognition. Function The helix-turn-helix motif is a DNA-binding motif. The recognition and binding to DNA by helix-turn-helix proteins is done by the two α helices, one occupying the N-terminus, N-terminal end of the mo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Leucine Zipper
A leucine zipper (or leucine scissors) is a common three-dimensional structural motif in proteins. They were first described by Landschulz and collaborators in 1988 when they found that an enhancer binding protein had a very characteristic 30-amino acid segment and the display of these amino acid sequences on an idealized alpha helix revealed a periodic repetition of leucine residues at every seventh position over a distance covering eight helical turns. The polypeptide segments containing these periodic arrays of leucine residues were proposed to exist in an alpha-helical conformation and the leucine side chains from one alpha helix interdigitate with those from the alpha helix of a second polypeptide, facilitating dimerization. Leucine zippers are a dimerization motif of the BZIP domain, bZIP (Basic-region leucine zipper) class of eukaryotic transcription factors. The bZIP domain is 60 to 80 amino acids in length with a highly conserved DNA binding basic region and a more divers ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
FokI
The restriction endonuclease Fok1, naturally found in ''Flavobacterium okeanokoites'', is a bacterial type IIS restriction endonuclease consisting of an N-terminal DNA-binding domain and a non sequence-specific DNA cleavage domain at the C-terminal. Once the protein is bound to duplex DNA via its DNA-binding domain at the 5'-GGATG-3' recognition site, the DNA cleavage domain is activated and cleaves the DNA at two locations, regardless of the nucleotide sequence at the cut site. The DNA is cut 9 nucleotides downstream of the motif on the forward strand, and 13 nucleotides downstream of the motif on the reverse strand, producing two sticky ends with 4-bp overhangs. Its molecular mass is 65.4 kDa, being composed of 587 amino acids. DNA-binding domain The recognition domain contains three subdomains (D1, D2 and D3) that are evolutionarily related to the DNA-binding domain of the catabolite gene activator protein which contains a helix-turn-helix. DNA-cleavage domain DNA cl ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Endonuclease
In molecular biology, endonucleases are enzymes that cleave the phosphodiester bond within a polynucleotide chain (namely DNA or RNA). Some, such as deoxyribonuclease I, cut DNA relatively nonspecifically (with regard to sequence), while many, typically called '' restriction endonucleases'' or ''restriction enzymes'', cleave only at very specific nucleotide sequences. Endonucleases differ from exonucleases, which cleave the ends of recognition sequences instead of the middle (''endo'') portion. Some enzymes known as "exo-endonucleases", however, are not limited to either nuclease function, displaying qualities that are both endo- and exo-like. Evidence suggests that endonuclease activity experiences a lag compared to exonuclease activity. Restriction enzymes are endonucleases from eubacteria and archaea that recognize a specific DNA sequence. The nucleotide sequence recognized for cleavage by a restriction enzyme is called the ''restriction site''. Typically, a restriction ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Position Identification With A Nuclease Tail
Proteins are large biomolecules and macromolecules that comprise one or more long chains of amino acid residues. Proteins perform a vast array of functions within organisms, including catalysing metabolic reactions, DNA replication, responding to stimuli, providing structure to cells and organisms, and transporting molecules from one location to another. Proteins differ from one another primarily in their sequence of amino acids, which is dictated by the nucleotide sequence of their genes, and which usually results in protein folding into a specific 3D structure that determines its activity. A linear chain of amino acid residues is called a polypeptide. A protein contains at least one long polypeptide. Short polypeptides, containing less than 20–30 residues, are rarely considered to be proteins and are commonly called peptides. The individual amino acid residues are bonded together by peptide bonds and adjacent amino acid residues. The sequence of amino acid residues i ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Z-DNA
Z-DNA is one of the many possible double helical structures of DNA. It is a left-handed double helical structure in which the helix winds to the left in a zigzag pattern, instead of to the right, like the more common B-DNA form. Z-DNA is thought to be one of three biologically active double-helical structures along with A-DNA and B-DNA. History Left-handed DNA was first proposed by Robert Wells and colleagues, as the structure of a repeating polymer of inosine–cytosine. They observed a "reverse" circular dichroism spectrum for such DNAs, and interpreted this incorrectly to mean that the strands wrapped around one another in a left-handed fashion. The relationship between Z-DNA and the more familiar B-DNA was indicated by the work of Pohl and Jovin, who showed that the ultraviolet circular dichroism of poly(dG-dC) was nearly inverted in 4 M sodium chloride solution and that the structure of poly d(I–C)·poly d(I–C) was in fact a right-handed D-DNA conformation. The ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chimera (protein)
Fusion proteins or chimeric (kī-ˈmir-ik) proteins (literally, made of parts from different sources) are proteins created through the joining of two or more genes that originally coded for separate proteins. Translation of this ''fusion gene'' results in a single or multiple polypeptides with functional properties derived from each of the original proteins. ''Recombinant fusion proteins'' are created artificially by recombinant DNA technology for use in biological research or therapeutics. '' Chimeric'' or ''chimera'' usually designate hybrid proteins made of polypeptides having different functions or physico-chemical patterns. ''Chimeric mutant proteins'' occur naturally when a complex mutation, such as a chromosomal translocation, tandem duplication, or retrotransposition creates a novel coding sequence containing parts of the coding sequences from two different genes. Naturally occurring fusion proteins are commonly found in cancer cells, where they may function as oncoprotei ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |