|
Binding Domain
In molecular biology, binding domain is a protein domain which binds to a specific atom or molecule, such as calcium or DNA. A protein domain is a part of a protein sequence and a tertiary structure that can change or evolve, function, and live by itself independent of the rest of the protein chain. Upon binding, proteins may undergo a conformational change. Binding domains are essential for the function of many proteins. They are essential because they help splice, assemble, and translate proteins.Yong, J., T. J. Golembe, D. J. Battle, L. Pellizzoni, and G. Dreyfuss. "SnRNAs Contain Specific SMN-binding Domains That Are Essential for SnRNP Assembly". ''Molecular and Cellular Biology''. U.S. National Library of Medicine, April 2004. Retrieved April 2017. Examples of binding domains include the Zinc finger, which binds to DNA, and EF hand, which binds to calcium. See also *DNA-binding domain *Receptor (biochemistry) In biochemistry and pharmacology, receptors are chemical ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Molecular Biology
Molecular biology is a branch of biology that seeks to understand the molecule, molecular basis of biological activity in and between Cell (biology), cells, including biomolecule, biomolecular synthesis, modification, mechanisms, and interactions. Though cells and other microscopic structures had been observed in living organisms as early as the 18th century, a detailed understanding of the mechanisms and interactions governing their behavior did not emerge until the 20th century, when technologies used in physics and chemistry had advanced sufficiently to permit their application in the biological sciences. The term 'molecular biology' was first used in 1945 by the English physicist William Astbury, who described it as an approach focused on discerning the underpinnings of biological phenomena—i.e. uncovering the physical and chemical structures and properties of biological molecules, as well as their interactions with other molecules and how these interactions explain observ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Domain
In molecular biology, a protein domain is a region of a protein's Peptide, polypeptide chain that is self-stabilizing and that Protein folding, folds independently from the rest. Each domain forms a compact folded Protein tertiary structure, three-dimensional structure. Many proteins consist of several domains, and a domain may appear in a variety of different proteins. Molecular evolution uses domains as building blocks and these may be recombined in different arrangements to create proteins with different functions. In general, domains vary in length from between about 50 amino acids up to 250 amino acids in length. The shortest domains, such as zinc fingers, are stabilized by metal ions or Disulfide bond, disulfide bridges. Domains often form functional units, such as the calcium-binding EF-hand, EF hand domain of calmodulin. Because they are independently stable, domains can be "swapped" by genetic engineering between one protein and another to make chimera (protein), chimeric ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Binding (molecular)
Molecular binding is an attractive interaction between two molecules that results in a stable association in which the molecules are in close proximity to each other. It is formed when atoms or molecules bind together by sharing of electrons. It often, but not always, involves some chemical bonding. In some cases, the associations can be quite strong—for example, the protein streptavidin and the vitamin biotin have a dissociation constant (reflecting the ratio between bound and free biotin) on the order of 10−14—and so the reactions are effectively irreversible. The result of molecular binding is sometimes the formation of a molecular complex in which the attractive forces holding the components together are generally non-covalent, and thus are normally energetically weaker than covalent bonds. Molecular binding occurs in biological complexes (e.g., between pairs or sets of proteins, or between a protein and a small molecule ligand it binds) and also in abiologic chemic ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Chemical Specificity
Chemical specificity is the ability of binding site of a macromolecule (such as a protein) to bind specific ligands. The fewer ligands a protein can bind, the greater its specificity. Specificity describes the strength of binding between a given protein and ligand. This relationship can be described by a dissociation constant, which characterizes the balance between bound and unbound states for the protein-ligand system. In the context of a single enzyme and a pair of binding molecules, the two ligands can be compared as stronger or weaker ligands (for the enzyme) on the basis of their dissociation constants. (A lower value corresponds to a stronger binding.) Specificity for a set of ligands is unrelated to the ability of an enzyme to catalyze a given reaction, with the ligand as a substrate. If a given enzyme has a high chemical specificity, this means that the set of ligands to which it binds is limited, such that neither binding events nor catalysis can occur at an apprecia ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Calcium
Calcium is a chemical element; it has symbol Ca and atomic number 20. As an alkaline earth metal, calcium is a reactive metal that forms a dark oxide-nitride layer when exposed to air. Its physical and chemical properties are most similar to its heavier homologues strontium and barium. It is the fifth most abundant element in Earth's crust, and the third most abundant metal, after iron and aluminium. The most common calcium compound on Earth is calcium carbonate, found in limestone and the fossils of early sea life; gypsum, anhydrite, fluorite, and apatite are also sources of calcium. The name comes from Latin ''calx'' " lime", which was obtained from heating limestone. Some calcium compounds were known to the ancients, though their chemistry was unknown until the seventeenth century. Pure calcium was isolated in 1808 via electrolysis of its oxide by Humphry Davy, who named the element. Calcium compounds are widely used in many industries: in foods and pharmaceuticals for ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Sequence
Protein primary structure is the linear sequence of amino acids in a peptide or protein. By convention, the primary structure of a protein is reported starting from the amino-terminal (N) end to the carboxyl-terminal (C) end. Protein biosynthesis is most commonly performed by ribosomes in cells. Peptides can also be synthesized in the laboratory. Protein primary structures can be directly sequenced, or inferred from DNA sequences. Formation Biological Amino acids are polymerised via peptide bonds to form a long backbone, with the different amino acid side chains protruding along it. In biological systems, proteins are produced during translation by a cell's ribosomes. Some organisms can also make short peptides by non-ribosomal peptide synthesis, which often use amino acids other than the encoded 22, and may be cyclised, modified and cross-linked. Chemical Peptides can be synthesised chemically via a range of laboratory methods. Chemical methods typically synt ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Tertiary Structure
Protein tertiary structure is the three-dimensional shape of a protein. The tertiary structure will have a single polypeptide chain "backbone" with one or more protein secondary structures, the protein domains. Amino acid side chains and the backbone may interact and bond in a number of ways. The interactions and bonds of side chains within a particular protein determine its tertiary structure. The protein tertiary structure is defined by its atomic coordinates. These coordinates may refer either to a protein domain or to the entire tertiary structure. A number of these structures may bind to each other, forming a quaternary structure. History The science of the tertiary structure of proteins has progressed from one of hypothesis to one of detailed definition. Although Emil Fischer had suggested proteins were made of polypeptide chains and amino acid side chains, it was Dorothy Maud Wrinch who incorporated geometry into the prediction of protein structures. Wrinch demonstr ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Evolution
Evolution is the change in the heritable Phenotypic trait, characteristics of biological populations over successive generations. It occurs when evolutionary processes such as natural selection and genetic drift act on genetic variation, resulting in certain characteristics becoming more or less common within a population over successive generations. The process of evolution has given rise to biodiversity at every level of biological organisation. The scientific theory of evolution by natural selection was conceived independently by two British naturalists, Charles Darwin and Alfred Russel Wallace, in the mid-19th century as an explanation for why organisms are adapted to their physical and biological environments. The theory was first set out in detail in Darwin's book ''On the Origin of Species''. Evolution by natural selection is established by observable facts about living organisms: (1) more offspring are often produced than can possibly survive; (2) phenotypic variatio ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Conformational Change
In biochemistry, a conformational change is a change in the shape of a macromolecule, often induced by environmental factors. A macromolecule is usually flexible and dynamic. Its shape can change in response to changes in its environment or other factors; each possible shape is called a conformation, and a transition between them is called a ''conformational change''. Factors that may induce such changes include temperature, pH, voltage, light in chromophores, concentration of ions, phosphorylation, or the binding of a ligand. Transitions between these states occur on a variety of length scales (tenths of Å to nm) and time scales (ns to s), and have been linked to functionally relevant phenomena such as allosteric signaling and enzyme catalysis. Laboratory analysis Many biophysical techniques such as crystallography, NMR, electron paramagnetic resonance (EPR) using spin label techniques, circular dichroism (CD), hydrogen exchange, and FRET can be used to study macromo ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Zinc Finger
A zinc finger is a small protein structural motif that is characterized by the coordination of one or more zinc ions (Zn2+) which stabilizes the fold. The term ''zinc finger'' was originally coined to describe the finger-like appearance of a hypothesized structure from the African clawed frog (''Xenopus laevis'') transcription factor IIIA. However, it has been found to encompass a wide variety of differing protein structures in eukaryotic cells. '' Xenopus laevis'' TFIIIA was originally demonstrated to contain zinc and require the metal for function in 1983, the first such reported zinc requirement for a gene regulatory protein followed soon thereafter by the Krüppel factor in ''Drosophila''. It often appears as a metal-binding domain in multi-domain proteins. Proteins that contain zinc fingers (zinc finger proteins) are classified into several different structural families. Unlike many other clearly defined supersecondary structures such as Greek keys or β hairpins, ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
EF Hand
The EF hand is a helix–loop–helix structural domain or ''motif'' found in a large family of calcium-binding proteins. The EF-hand motif contains a helix–loop–helix topology, much like the spread thumb and forefinger of the human hand, in which the Ca2+ ions are coordinated by ligands within the loop. The motif takes its name from traditional nomenclature used in describing the protein parvalbumin, which contains three such motifs and is probably involved in muscle relaxation via its calcium-binding activity. The EF-hand consists of two alpha helices linked by a short loop region (usually about 12 amino acids) that usually binds calcium ions. EF-hands also appear in each structural domain of the signaling protein calmodulin and in the muscle protein troponin-C. Calcium ion binding site The calcium ion is coordinated in a pentagonal bipyramidal configuration. The six residues involved in the binding are in positions 1, 3, 5, 7, 9 and 12; these residues are denoted ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
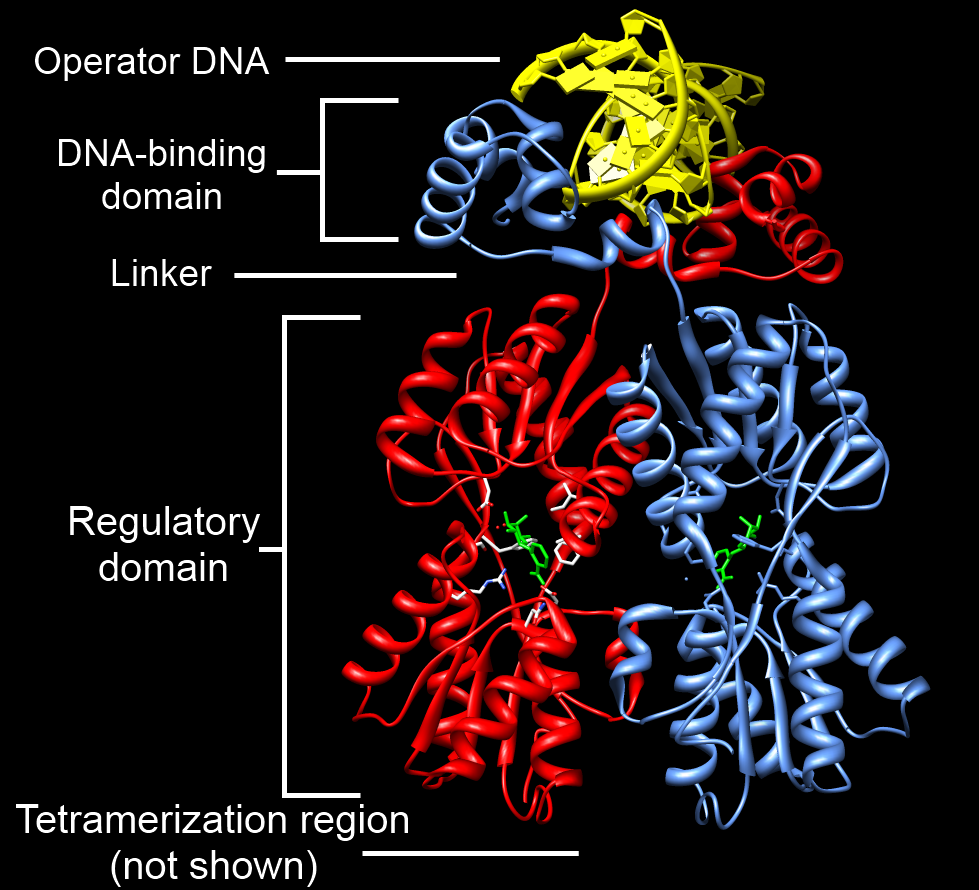
DNA-binding Domain
A DNA-binding domain (DBD) is an independently folded protein domain that contains at least one structural motif that recognizes double- or single-stranded DNA. A DBD can recognize a specific DNA sequence (a recognition sequence) or have a general affinity to DNA. Some DNA-binding domains may also include nucleic acids in their folded structure. Function One or more DNA-binding domains are often part of a larger protein consisting of further protein domains with differing function. The extra domains often regulate the activity of the DNA-binding domain. The function of DNA binding is either structural or involves transcription regulation, with the two roles sometimes overlapping. DNA-binding domains with functions involving DNA structure have biological roles in DNA replication, repair, storage, and modification, such as methylation. Many proteins involved in the regulation of gene expression contain DNA-binding domains. For example, proteins that regulate transcription ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |