|
ATPase Inhibitors
ATPases (, Adenosine 5'-TriPhosphatase, adenylpyrophosphatase, ATP monophosphatase, triphosphatase, SV40 T-antigen, ATP hydrolase, complex V (mitochondrial electron transport), (Ca2+ + Mg2+)-ATPase, HCO3−-ATPase, adenosine triphosphatase) are a class of enzymes that catalyze the decomposition of ATP into ADP and a free phosphate ion or the inverse reaction. This dephosphorylation reaction releases energy, which the enzyme (in most cases) harnesses to drive other chemical reactions that would not otherwise occur. This process is widely used in all known forms of life. Some such enzymes are integral membrane proteins (anchored within biological membranes), and move solutes across the membrane, typically against their concentration gradient. These are called transmembrane ATPases. Functions Transmembrane ATPases import metabolites necessary for cell metabolism and export toxins, wastes, and solutes that can hinder cellular processes. An important example is the sodium-potas ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Cell (biology)
The cell is the basic structural and functional unit of life forms. Every cell consists of a cytoplasm enclosed within a membrane, and contains many biomolecules such as proteins, DNA and RNA, as well as many small molecules of nutrients and metabolites.Cell Movements and the Shaping of the Vertebrate Body in Chapter 21 of Molecular Biology of the Cell '' fourth edition, edited by Bruce Alberts (2002) published by Garland Science. The Alberts text discusses how the "cellular building blocks" move to shape developing embryos. It is also common to describe small molecules such as ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Oxidative Phosphorylation
Oxidative phosphorylation (UK , US ) or electron transport-linked phosphorylation or terminal oxidation is the metabolic pathway in which cells use enzymes to oxidize nutrients, thereby releasing chemical energy in order to produce adenosine triphosphate (ATP). In eukaryotes, this takes place inside mitochondria. Almost all aerobic organisms carry out oxidative phosphorylation. This pathway is so pervasive because it releases more energy than alternative fermentation processes such as anaerobic glycolysis. The energy stored in the chemical bonds of glucose is released by the cell in the citric acid cycle producing carbon dioxide, and the energetic electron donors NADH and FADH. Oxidative phosphorylation uses these molecules and O2 to produce ATP, which is used throughout the cell whenever energy is needed. During oxidative phosphorylation, electrons are transferred from the electron donors to a series of electron acceptors in a series of redox reactions ending in oxyg ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Active Transport
In cellular biology, ''active transport'' is the movement of molecules or ions across a cell membrane from a region of lower concentration to a region of higher concentration—against the concentration gradient. Active transport requires cellular energy to achieve this movement. There are two types of active transport: primary active transport that uses adenosine triphosphate ( ATP), and secondary active transport that uses an electrochemical gradient. Some examples of active transport include: * Phagocytosis of bacteria by macrophages * Movement of calcium ions out of cardiac muscle cells * Transportation of amino acids across the intestinal lining in the human gut * Secretion of proteins such as enzymes, peptide hormones, and antibodies from various cells * Functioning of white blood cells to defend invading diseases Active cellular transportation (ACT) Unlike passive transport, which uses the kinetic energy and natural entropy of molecules moving down a gradient, activ ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Protein Design
Protein design is the rational design of new protein molecules to design novel activity, behavior, or purpose, and to advance basic understanding of protein function. Proteins can be designed from scratch (''de novo'' design) or by making calculated variants of a known protein structure and its sequence (termed ''protein redesign''). Rational protein design approaches make protein-sequence predictions that will fold to specific structures. These predicted sequences can then be validated experimentally through methods such as peptide synthesis, site-directed mutagenesis, or artificial gene synthesis. Rational protein design dates back to the mid-1970s. Recently, however, there were numerous examples of successful rational design of water-soluble and even transmembrane peptides and proteins, in part due to a better understanding of different factors contributing to protein structure stability and development of better computational methods. Overview and history The goal in rat ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Nest (protein Structural Motif)
The Nest is a type of protein structural motif. It is a small recurring anion-binding feature of both proteins and peptides. Each consists of the main chain atoms of three consecutive amino acid residues. The main chain NH groups bind the anions while the side chain atoms are often not involved. Proline residues lack NH groups so are rare in nests. About one in 12 of amino acid residues in proteins, on average, belongs to a nest. Nest conformations The conformation of a nest is such that the NH groups of the first and third amino acid residues are liable to be hydrogen bonded to a negatively charged, or partially negatively charged, atom, often an oxygen atom. The NH of the second residue may also be hydrogen bonded to the same atom but usually points somewhat away. These main chain atoms form a concavity called a nest into which an anionic atom fits. Such anionic atoms are sometimes called eggs and more than one egg may occur bound to a nest. The oxyanion hole of the intesti ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Alpha Helix
The alpha helix (α-helix) is a common motif in the secondary structure of proteins and is a right hand- helix conformation in which every backbone N−H group hydrogen bonds to the backbone C=O group of the amino acid located four residues earlier along the protein sequence. The alpha helix is also called a classic Pauling–Corey–Branson α-helix. The name 3.613-helix is also used for this type of helix, denoting the average number of residues per helical turn, with 13 atoms being involved in the ring formed by the hydrogen bond. Among types of local structure in proteins, the α-helix is the most extreme and the most predictable from sequence, as well as the most prevalent. Discovery In the early 1930s, William Astbury showed that there were drastic changes in the X-ray fiber diffraction of moist wool or hair fibers upon significant stretching. The data suggested that the unstretched fibers had a coiled molecular structure with a characteristic repeat of ≈. ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Beta Sheet
The beta sheet, (β-sheet) (also β-pleated sheet) is a common motif of the regular protein secondary structure. Beta sheets consist of beta strands (β-strands) connected laterally by at least two or three backbone hydrogen bonds, forming a generally twisted, pleated sheet. A β-strand is a stretch of polypeptide chain typically 3 to 10 amino acids long with backbone in an extended conformation. The supramolecular association of β-sheets has been implicated in the formation of the fibrils and protein aggregates observed in amyloidosis, notably Alzheimer's disease. History The first β-sheet structure was proposed by William Astbury in the 1930s. He proposed the idea of hydrogen bonding between the peptide bonds of parallel or antiparallel extended β-strands. However, Astbury did not have the necessary data on the bond geometry of the amino acids in order to build accurate models, especially since he did not then know that the peptide bond was planar. A refined version w ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
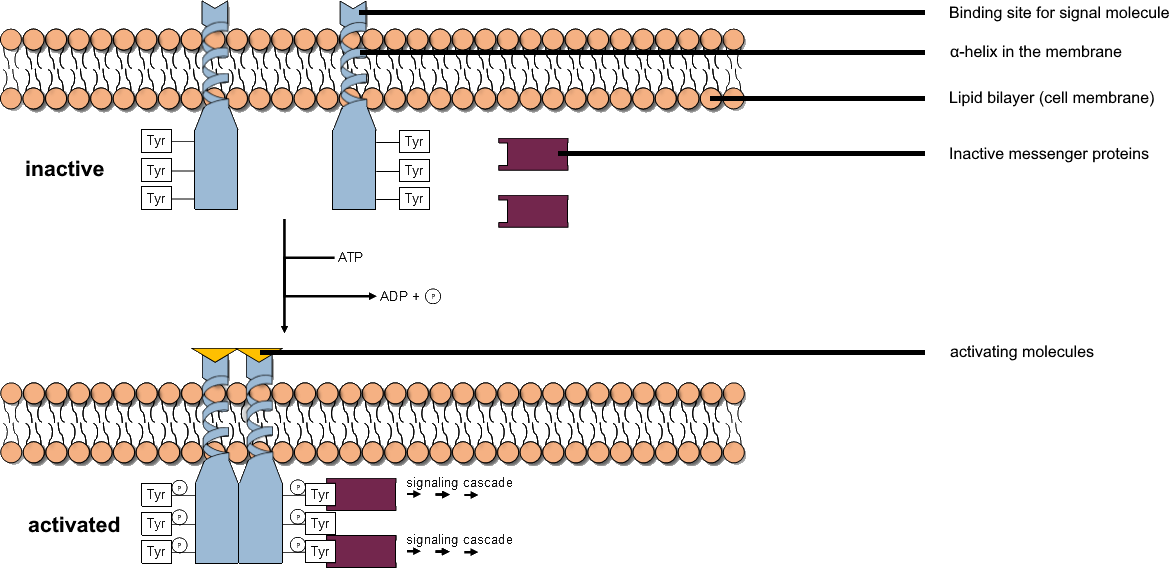
Tyrosine Kinase
A tyrosine kinase is an enzyme that can transfer a phosphate group from ATP to the tyrosine residues of specific proteins inside a cell. It functions as an "on" or "off" switch in many cellular functions. Tyrosine kinases belong to a larger class of enzymes known as protein kinases which also attach phosphates to other amino acids such as serine and threonine. Phosphorylation of proteins by kinases is an important mechanism for communicating signals within a cell (signal transduction) and regulating cellular activity, such as cell division. Protein kinases can become mutated, stuck in the "on" position, and cause unregulated growth of the cell, which is a necessary step for the development of cancer. Therefore, kinase inhibitors, such as imatinib and osimertinib, are often effective cancer treatments. Most tyrosine kinases have an associated protein tyrosine phosphatase, which removes the phosphate group. Reaction Protein kinases are a group of enzymes that possess a c ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Walker Motifs
The Walker A and Walker B motifs are protein sequence motifs, known to have highly conserved three-dimensional structures. These were first reported in ATP-binding proteins by Walker and co-workers in 1982. Of the two motifs, the A motif is the main "P-loop" responsible for binding phosphate, while the B motif is a much less conserved downstream region. The P-loop is best known for its presence in ATP- and GTP-binding proteins, and is also found in a variety of proteins with phosphorylated substrates. Major lineages include: * RecA and rotor ATP synthase / ATPases (α and β subunits). * Nucleic acid-dependent ATPases: helicases, Swi2, and PhoH () * AAA proteins * STAND NTPases including MJ, PH, AP, and NACHT ATPases * ABC- PilT ATPases * Nucleotide kinases () * G domain proteins: G-proteins (transducin), myosin. Walker A motif Walker A motif, also known as the Walker loop, or P-loop, or phosphate-binding loop, is a motif in proteins that is associated with phosphate bi ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Co-transport
In cellular biology, ''active transport'' is the movement of molecules or ions across a cell membrane from a region of lower concentration to a region of higher concentration—against the concentration gradient. Active transport requires cellular energy to achieve this movement. There are two types of active transport: primary active transport that uses adenosine triphosphate ( ATP), and secondary active transport that uses an electrochemical gradient. Some examples of active transport include: * Phagocytosis of bacteria by macrophages * Movement of calcium ions out of cardiac muscle cells * Transportation of amino acids across the intestinal lining in the human gut * Secretion of proteins such as enzymes, peptide hormones, and antibodies from various cells * Functioning of white blood cells to defend invading diseases Active cellular transportation (ACT) Unlike passive transport, which uses the kinetic energy and natural entropy of molecules moving down a gradient, active ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |
Hydrogen Potassium ATPase
Gastric hydrogen potassium ATPase, also known as H+/K+ ATPase, is an enzyme which functions to acidify the stomach. It is a member of the P-type ATPases, also known as E1-E2 ATPases due to its two states. Biological function and location The gastric hydrogen potassium ATPase or H+/K+ ATPase is the proton pump of the stomach. It exchanges potassium from the intestinal lumen with cytoplasmic hydronium and is the enzyme primarily responsible for the acidification of the stomach contents and the activation of the digestive enzyme pepsin (see gastric acid). The H+/K+ ATPase is found in parietal cells, which are highly specialized epithelial cells located in the inner cell lining of the stomach called the gastric mucosa. Parietal cells possess an extensive secretory membrane system and the H+/K+ ATPase is the major protein constituent of these membranes. A small amount of H+/K+ ATPase is also found in the renal medulla. Genes and protein structure The H+/K+ ATPase is a heterod ... [...More Info...] [...Related Items...] OR: [Wikipedia] [Google] [Baidu] |